Figure 1.
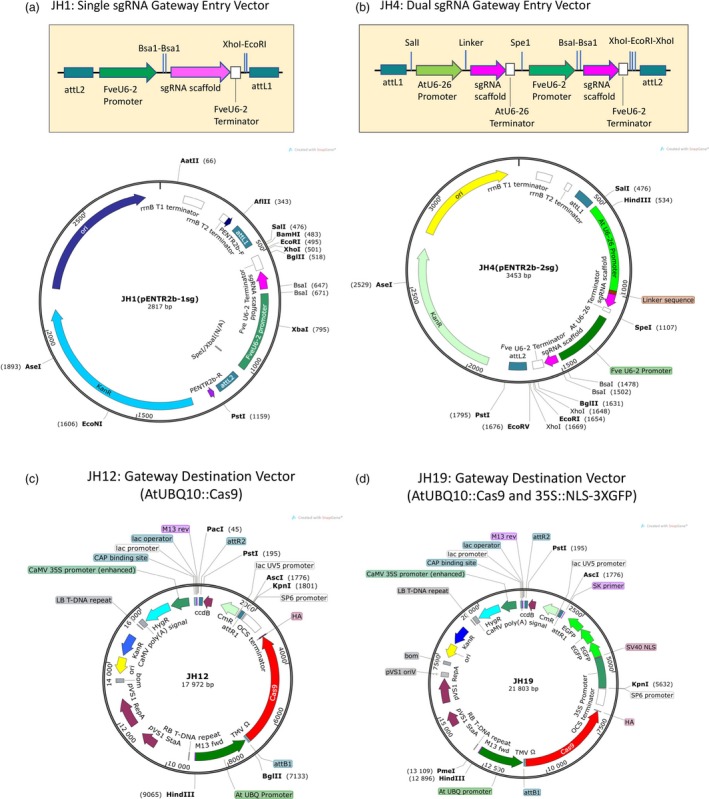
Diagrams illustrating gateway‐based CRISPR vectors. (a) Illustration of the single sgRNA gateway entry vector JH1. A sgRNA cloning cassette (enlarged on top) is inserted between the attL1 and attL2 recombination sites of pENTR2b. Two inverted BsaI restriction enzyme recognition sites enable easy cloning of the double‐stranded target sequence. pENTR2b‐R and pENTR2b‐F in the vector map are two sequencing primers. (b) Illustration of the dual sgRNA gateway entry vector JH4. The dual sgRNA cloning cassette (enlarged on top) is inserted between the attL1 and attL2 recombination sites of pENTR2b. One sgRNA is driven by the FveU6‐2 promoter similarly to JH1. The second sgRNA is driven by the Arabidopsis U6‐26 promoter. A linker sequence GCGCTTCAAGGTGCACATGG between the AtU6‐26 promoter and the sgRNA scaffold can be replaced with a 20 bp target sequence using the Q5 site‐directed mutagenesis kit. (c) The gateway destination vector JH12 is a hygromycin‐resistant binary vector harbouring a AtUBQ10 promoter‐driven Cas9. (d) The gateway destination vector JH19, a binary vector similar to JH12 except that JH19, harbours a 35S::NSL‐3XGFP cassette. The GFP fluorescence allows easy visualization and identification of transgenic plants.
