Figure 5.
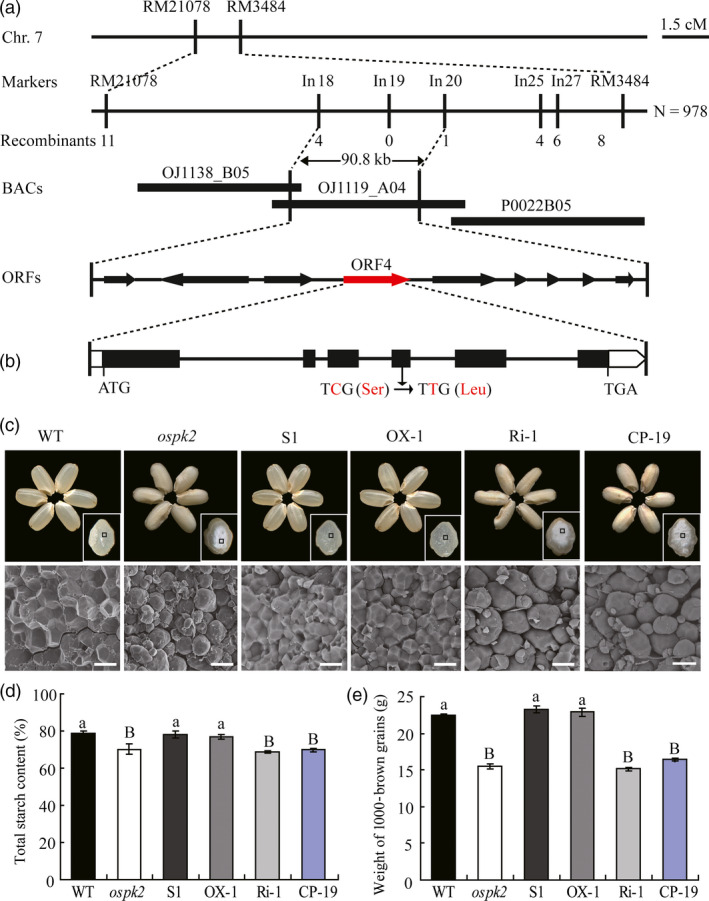
Map‐based cloning and complementation of the ospk2 mutant. (a) Fine mapping of the OsPK2 locus. The OsPK2 locus was mapped to a 90.8 kb region by markers In18 and In20 on chromosome 7 (Chr.7) which contained nine predicted genes. The molecular markers and the number of recombinants are shown. cM, centimorgan; ORF, open reading frame. (b) Gene structure and mutation site in OsPK2. Base pair change (C to T) detected in ospk2 in the 4th exon of Os07g0181000 causing a Ser‐296 to Leu‐296 change. ATG and TGA represent the start and stop codons, respectively. (c) Complementation of the ospk2 mutation in transgenic lines (S1) and overexpression of OsPK2 in ospk2 (OX‐1) showing the restored wild‐type seed appearance and normal starch granules in endosperm, whereas RNAi (Ri‐1) and CRISPR/Cas9 mediated editing (CP‐19) of OsPK2 in ZH11 background produce abnormal seed appearance and seeds became chalky. Bars: 20 μm. (d, e) Total starch content and 1000‐brown grains weight of grains in complementation (S1), overexpression (OX‐1), RNAi (Ri‐1) and CRISPR/Cas9 mediated editing lines (CP‐19). Data are shown as means ± SD from three biological replicates, each replication in (e) is not less than 200 seeds. Significant difference analysed by multiple‐comparison. Different letters indicate statistically significant difference (small letter, P < 0.05 and capital letter, P < 0.01).
