Figure 6.
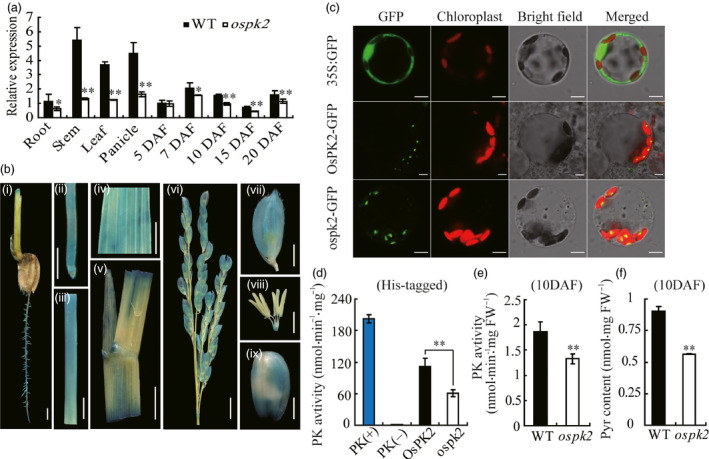
Expression pattern, subcellular localization and enzyme assay of OsPK2. (a) OsPK2 expression level in various tissues and in developing endosperms of the WT and ospk2. RNA was isolated from root, stem, leaf and panicle at the heading stage and developing endosperms at 5, 7, 10, 15 and 20 days after fertilization (DAF). Values are means ± SD from three biological replicates. Asterisks indicate statistical significance between wild‐type and mutant, as determined by a Student's t‐test (*P < 0.05, **P < 0.01). (b) GUS staining in root (i, ii), stem (iii), leaf (iv), leaf sheath (v), panicle (vi), glume (vii), spikelet (viii) and brown rice (ix) driven by the OsPK2 promoter. Bar: 2 mm (i, vii–ix); 1 cm (ii–v,); 5 mm (vi). (c) Subcellular localization of OsPK2 in rice protoplast cells. From the top panel to the bottom panel: free GFP used as control; OsPK2 full‐length coding region and GFP fusion protein (OsPK2‐GFP) and ospk2 full‐length coding region and GFP fusion protein (ospk2‐GFP). Forty‐eight hours after transformation, protoplast cells were observed using a confocal laser scanning microscope. GFP signals, chlorophyll autofluorescence, bright‐field images and the merged images of GFP and chlorophyll signals are shown in each panel. Bars: 5 μm. (d) Enzyme activity assay of OsPK2 and ospk2 protein expressed in baculovirus system and purified by His‐tag. PK activity was determined by the decreasing of NADH (measured the absorbance value at 340 nm for 2 min). (e) PK activity of fresh seeds (10 DAF) from wild‐type and ospk2. (f) Pyruvate content of seeds (10 DAF) from wild‐type and ospk2. Data in (d–f) are shown as mean ± SD from three biological replicates. Asterisks indicate statistical significance as determined by a Student's t‐test (*P < 0.05, **P < 0.01).
