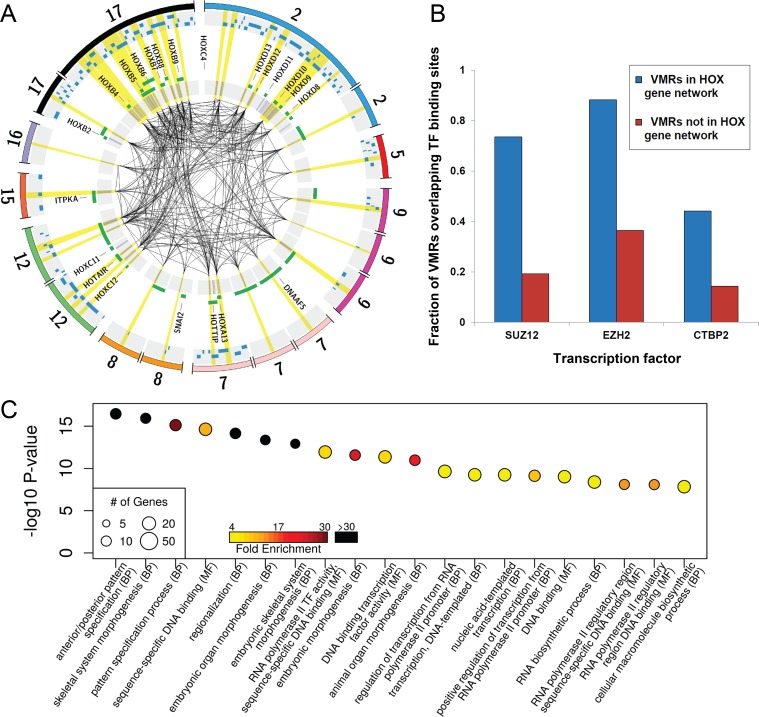
Fig 4. Cis and trans co-regulation of VMRs located at functionally related networks of genes that govern key developmental pathways.
(A) After selecting one CpG per VMR with the highest variance, we applied WGCNA to identify networks of significantly co-regulated VMRs. The Circos plot shows a representation of one of the largest co-regulated VMR modules identified in fibroblasts, which comprises 34 independent VMRs located on four different chromosomes, comprising all four clusters of HOX genes (outer circle). CpGs within VMRs in the co-regulated module are represented by blue tick marks (inner grey circle), with black lines joining VMRs that have methylation levels with pair wise absolute correlation values R≥0.7 (highlighted in yellow). Green bars show locations of genes at each locus. Blue bars show the location of transcription factor binding sites for SUZ12, EZH2 and CTBP2, all of which are significantly enriched within this co-regulated module. (B) Analysis of transcription factor binding sites defined using ChIP-seq [64] showed that VMRs within the co-regulated HOX gene module shown in (A) are significantly enriched for SUZ12, EZH2 and CTBP2 binding compared to all VMRs defined in fibroblasts (Bonferroni corrected p = 5.3x10-11, p = 1.5x10-9 and p = 5x10-5, respectively). Thus, binding of these TFs represents a potential mechanism by which epigenetic variation could be coordinated at multiple independent loci in trans. (C) Results of Gene Ontology (GO) analysis of genes associated with VMRs in the most significant co-regulated module identified in fibroblasts. We identified highly significant enrichments for multiple biological processes, including body patterning, growth and morphogenesis (S5 Table).