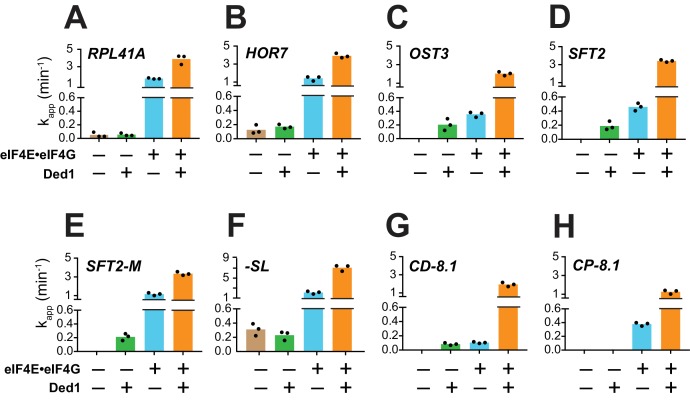
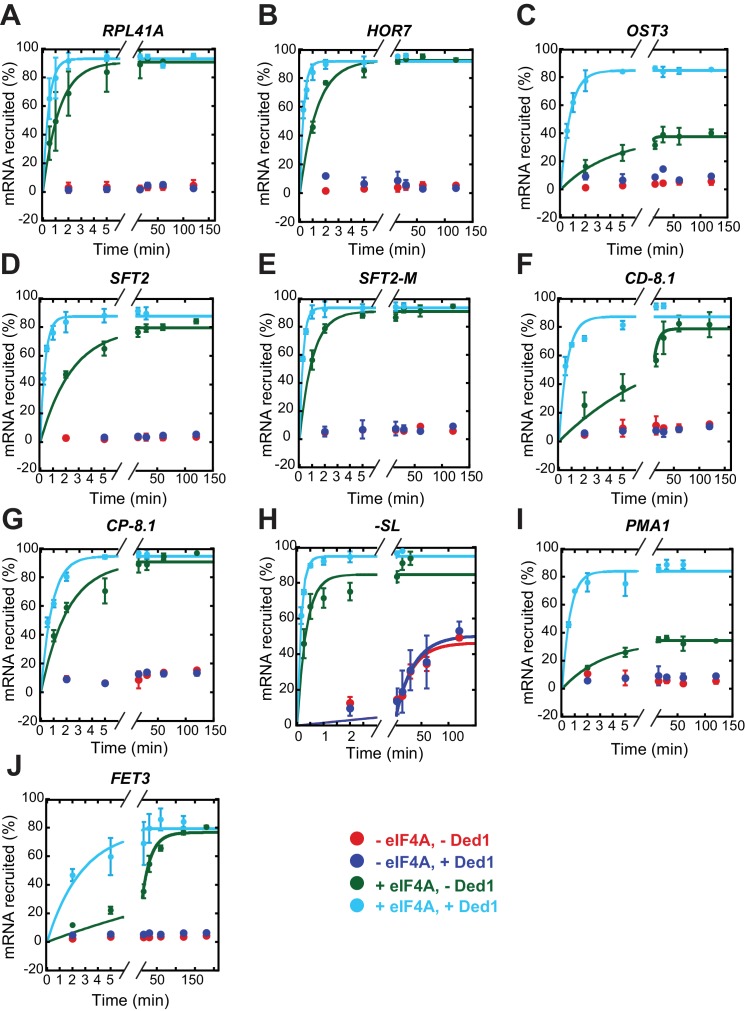
Figure 3. Ded1 acceleration of 48S PIC assembly is completely dependent on eIF4E·eIF4G for a subset of mRNAs.
(A–H) Apparent rates (kapp) of 48S PIC assembly for the indicated mRNAs observed without eIF4E·eIF4G and Ded1 (tan bars), with Ded1 but no eIF4E·eIF4G (green bars), with eIF4E·eIF4G but no Ded1 (blue bars), or with both Ded1 and eIF4E·eIF4G (orange bars). Note the different Y-axis scale for -SL (F). See Figure 3—figure supplement 1 and Figure 3—source data 1.