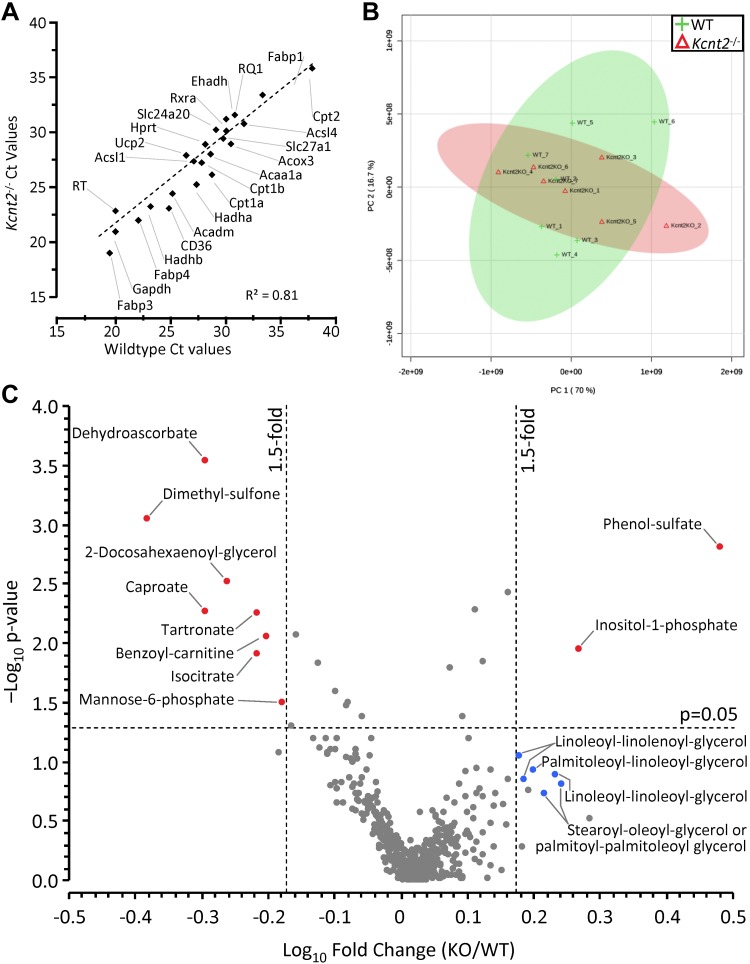
Figure 8.
Kcnt2−/− cardiac expression profiling and metabolomics. A) qPCR Ct values for 27 metabolically important genes (Table 1) in WT and Kcnt2−/− hearts (n = 3 independent RNA preparations/genotype). Data are mean, errors are omitted for clarity. B) WT and Kcnt2−/− hearts were perfused with KH buffer containing palmitate+glucose and freeze clamped for metabolomic analysis by liquid chromatography-MS/MS. Graph shows principle component analysis of 501 cardiac metabolites. The first and second principal components contributed 86.7% of the overall metabolic character. Shaded ovals overlaying the graph indicate 95% confidence intervals for WT (lime) and Kcnt2−/− (salmon pink) samples. C) Volcano plot of the metabolic profile of Kcnt2−/− vs. WT hearts. Dashed lines: P = 0.05 cut off (y axis) and 1.5-fold change cut off (x axis). Each point represents a single metabolite, and each point represents means (n = 7 mice). Errors are omitted for clarity. Red: metabolites meeting fold-change and P-value criteria; blue: additional metabolites discussed in the text. Diamonds: males; circles: data from both sexes combined.