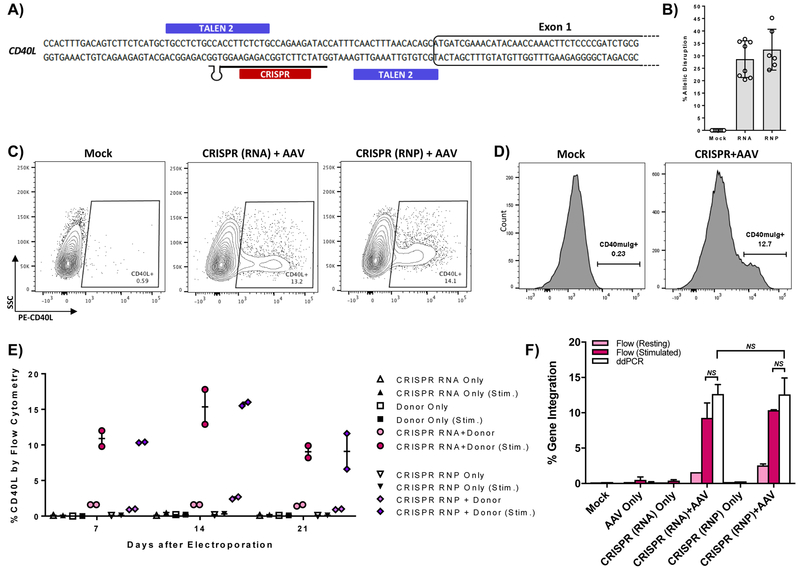
Figure 2. CRISPR/Cas9 mediated gene editing of the CD40L gene in XHIM T cells.
A) CRISPR gRNA binding site in the 5’UTR of the CD40L gene. B) Allelic disruption rates in XHIM CD4+ T cells using gRNA with Cas9 mRNA or complexed to recombinant Cas9 protein as RNP were measured by Surveyor nuclease assay (n=6–10 biological replicates in primary T cells from 4 different XHIM patients). C) Representative flow cytometry plots of gene-modified XHIM T cells after stimulation with anti-CD3/CD28 beads. D) CD40-muIg binding by CRISPR/Cas9 modified XHIM T cells to assess CD40L protein function. E) Expression of CD40L by flow cytometry in XHIM T cells treated with CRISPR/Cas9 RNA or RNP and AAV donor and re-stimulated over time in culture. F) Average gene integration rates in primary XHIM T cells measured by flow cytometry and ddPCR (n=2–4 biological replicates, 2 XHIM T cell donors). ddPCR was designed with a forward primer upstream of the 5’ homology arm, a reverse primer spanning exons 1 & 2, and a probe specific for the codon-optimized exon 1. Data in F were analyzed by Wilcoxon Rank-Sum Test. NS = not significant. See also Figure S3.