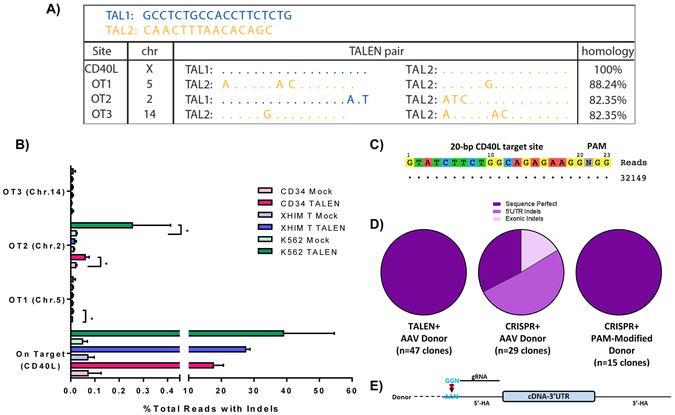
Figure 6. Off-Target Analysis and Characterization of HDR-Mediated Junctions.
A) Off-target sites as identified by GFP-IDLV trapping in TALEN-treated K562 cells. Allowing for homodimerization of TALEN arms, degree of homology to the on-target binding site is illustrated. B) Targeted re-sequencing of on-target and off-target sites by high-throughput sequencing in PBSC, XHIM CD4+ T cells, and K562 cells treated with TALEN mRNA (PBSC, XHIM T cell) or expression plasmids (K562). Data were analyzed by Wilcoxon Rank-Sum Test. *p≤0.05. C) Off target analysis of CRISPR 3 by GUIDE-seq in K562 cells treated with CRISPR RNP and double-stranded oligodeoxynucleotides. D) Characterization of HDR-mediated junctions between the endogenous CD40L genomic locus and the 5’ end of the integrated donor cassette in samples treated with either TALEN or CRISPR and the codon-optimized donor. Samples treated with CRISPRs required a donor containing a PAM site modification depicted in E) to prevent re-binding of the CRISPR gRNA causing unwanted repeat cleavage after cDNA integration. See also Figure S6.