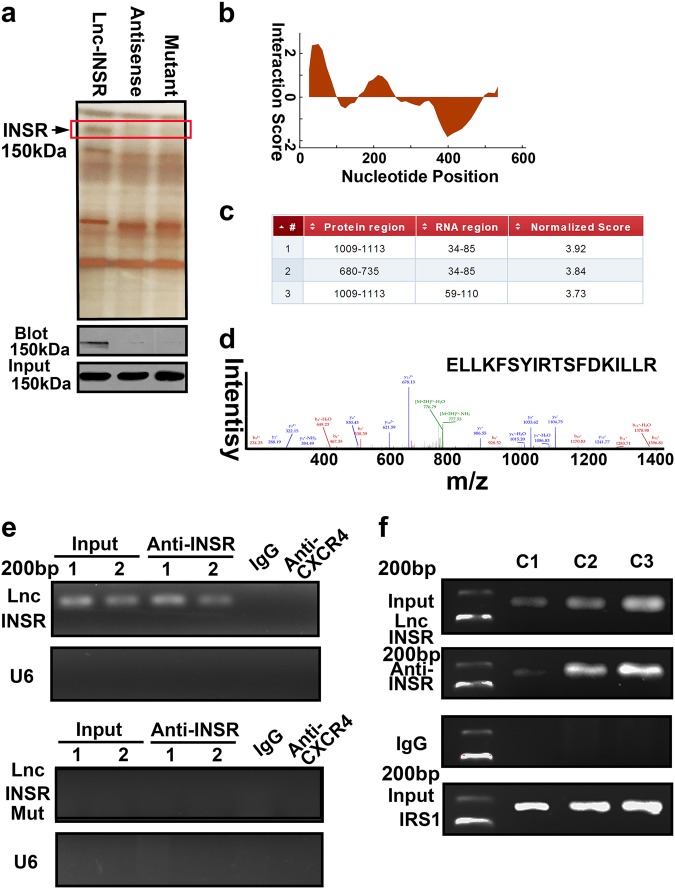
Fig. 4. Lnc-INSR direct binding with INSR at 1009–1113aa.
a RNA pull-down experiment with CD4 (+) T cell cytoplasmic extract in different groups was presented as silver staining. Specific bands were identified by immunoblot of INSR. b The predicted binding site for lnc-INSR with INSR analyzed by CatRAPID. c Detailed binding site located in the RNA sequence of lnc-INSR and protein residue of INSR. d Mass spectrometry identification of special amino acid of INSR. e RIP assay was performed using INSR antibody and was validated by agarose electrophoresis by using different primer. CXCR4 antibody and U6 primer was used as negative control. f Cells were treated with different concentration of lnc-INSR lentivirus (C1 indicate 1U, C2 indicated 2U, and C3 indicated 3U comparing with the basal 20 TU/ml according to manufacturer). IRS1 was used as positive controls