Fig. 6.
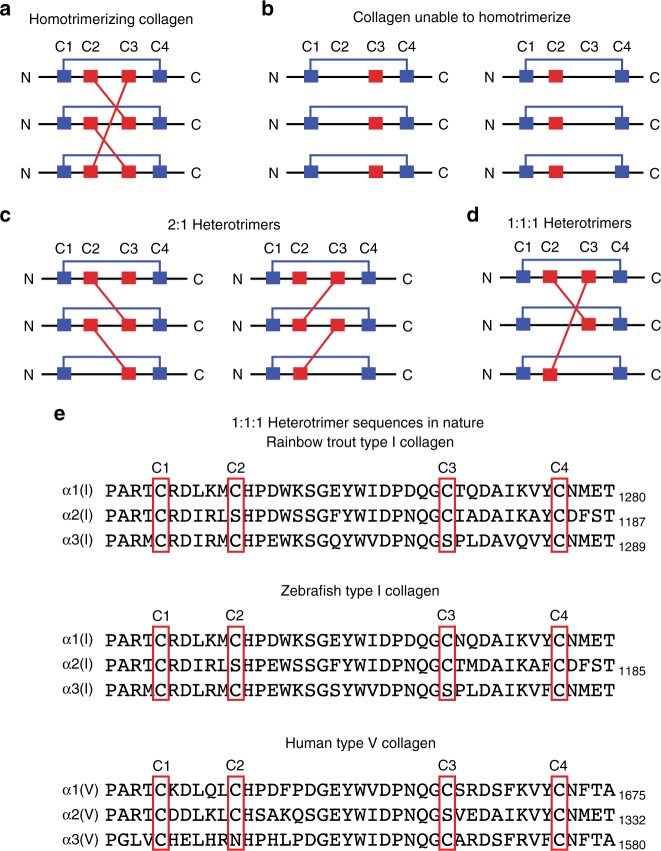
A priori prediction of collagen trimerization propensities based on the C2/C3 pattern (C1 through C4 shown). a Known disulfide-bonding network for a homotrimeric collagen strand with C2 and C3 intact, as revealed by the crystal structures of C-Proα1(I) and C-Proα1(III) homotrimers16, 17. b Predicted disulfide-bonding network for a C-Pro domain lacking a cysteine residue in position 2 or 3. Although intrastrand disulfide bonds likely form, interstrand disulfide bonds cannot form in the absence of C2 or C3. c Predicted disulfide-bonding network for a collagen heterotrimer that requires 2:1 assembly. The accuracy of this predicted disulfide-bonding pattern in 2:1 heterotrimers is supported by previous work modeling the replacement of a single C-Proα1(I) subunit in the homotrimer crystal structure with a C-Proα2(I) monomer16. Note the single free cysteine in the assembled protein owing to the presence of an odd number of cysteine residues. d Predicted disulfide-bonding network for a collagen heterotrimer that requires 1:1:1 assembly, illustrating the minimum requirements for stable 1:1:1 heterotrimer formation, which are one strand having cysteine residues at positions 2 and 3, one only at position 2, and one only at position 3. e Alignment of rainbow trout and zebrafish collagen-I C-Pro domains (C1–C4 only shown). The expected cysteine pattern for a 1:1:1 homotrimer in d is present in both instances where collagen-I has three α strands in nature and in human collagen type-V, which all form 1:1:1 heterotrimers. Note that in all images the potentially interstrand disulfide-forming cysteines (C2 and C3) are colored red, while the intrastrand disulfide-forming cysteines are colored blue