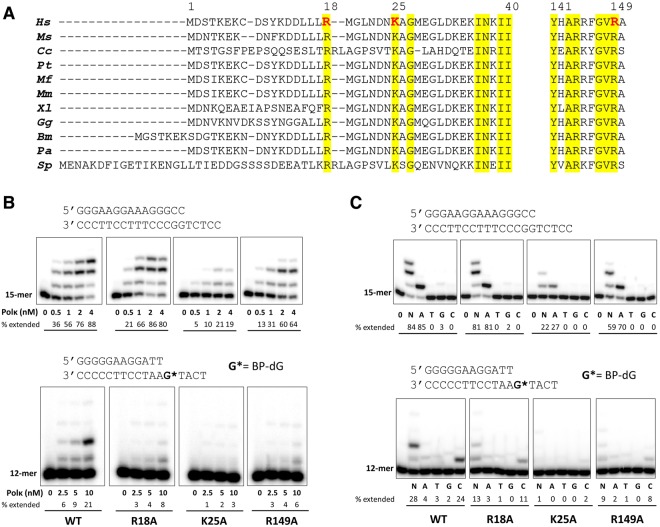
Figure 2.
Comparison of polκ from various eukaryotes, and extension of 32P-labeled primers by wild-type and mutant human polκ on normal and BP-dG-containing template. (A) Amino acid sequence alignments of the N-terminal extension and finger domains of polκ from various eukaryotes. Hs: Homo sapiens; Ms: mouse (Mus musculus); Cc: Coprinopsis cinereal; Pt: Pan troglodytes; Mf: Macaca fascicularis; Mm: Macaca mulatta; Xl: Xenopus laevis; Gg: Gallus gallus; Bm: Bos mutus; Pa: Pteropus alecto; Sp: Schizosaccharomyces pombe. Amino acid numbering corresponds to human polκ. Conserved residues are highlighted in yellow, and polκ residues analyzed in this study are red. (B) Reactions were done using 50 nM DNA substrate (primer/template) with increasing concentrations of wild-type and mutant human polκ (0–4 nM for normal DNA and 0–10 nM for BP-dG-containing DNA). The 32P-labeled 15-mer (for normal DNA) and 12-mer (for BP-dG DNA) primer was extended in the presence of all four dNTPs for 5 minutes for normal DNA (upper panel) and 10 minutes for BP-dG-containing DNA (lower panel) at 37 °C. (C) Reactions were carried out with no dNTPs (0) or a single dNTP type (A, T, G, or C) or a mixture of all four dNTPs (N) using 50 nM DNA substrate and 2 nM polκ for normal DNA (upper panel) and 10 nM polκ for BP-dG-containing DNA (lower panel). The reactions times were the same as in (B). Cropped images in each panel (upper or lower in B or C) are a composite of lanes from the same polyacrylamide gels. Brightness and contrasts were adjusted to improve the clarity. Original (unmodified images) representing (B) and (C) are included in Supplementary Figs S4 and S5, respectively. The sequences of DNA used in the assays are shown above the respective image panels. Percentage of each primer extended in the presence of a correct dNTP or mixed four nucleotides was quantified and is indicated below each lane.