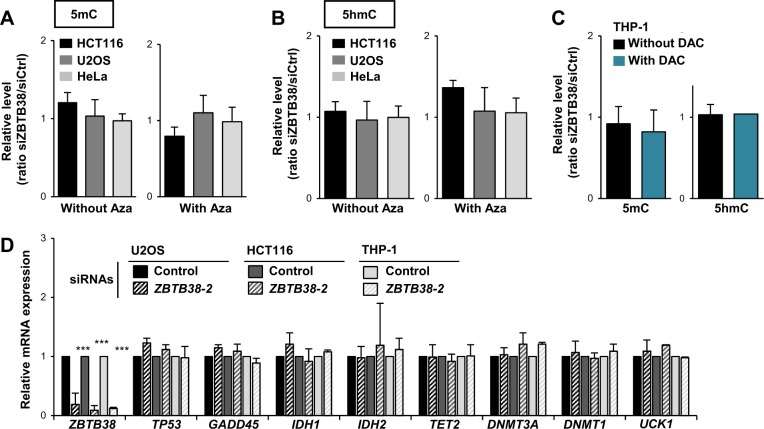
Fig. 2. Inactivation of ZBTB38 marginally impacts DNA demethylation and 5-azacytidine induced DNA demethylation.
a Dot blot analysis of CpG methylation levels in HCT116 (black bar), U2OS (grey bar), and HeLa (light grey bar) cells treated with siRNA against ZBTB38 or control siRNA and further exposed to 5-azacytidine (n = 3). The data represent relative levels of the condition siRNA ZBTB38 versus siRNA control. b Dot blot analysis of CpG hydroxymethylation levels in HCT116 (black bar), U2OS (grey bar), and HeLa (light grey bar) cells treated with siRNA against ZBTB38 or control siRNA and further exposed to 5-azacytidine (n = 3). The data represent relative levels of the condition siRNA ZBTB38 versus siRNA control. c Dot blot analysis of CpG methylation and hydroxymethylation levels in THP-1 cells treated with siRNA against ZBTB38 or control siRNA and further exposed to decitabine (n = 2). The data represent relative levels of the condition siRNA ZBTB38 versus siRNA control. d RT-PCR analysis of genes modulating 5-azacytidine toxicity in HCT116, U2OS, and THP-1 cells treated with siRNA against ZBTB38 (dashed bars) and control siRNA (solid bars) (n = 3)