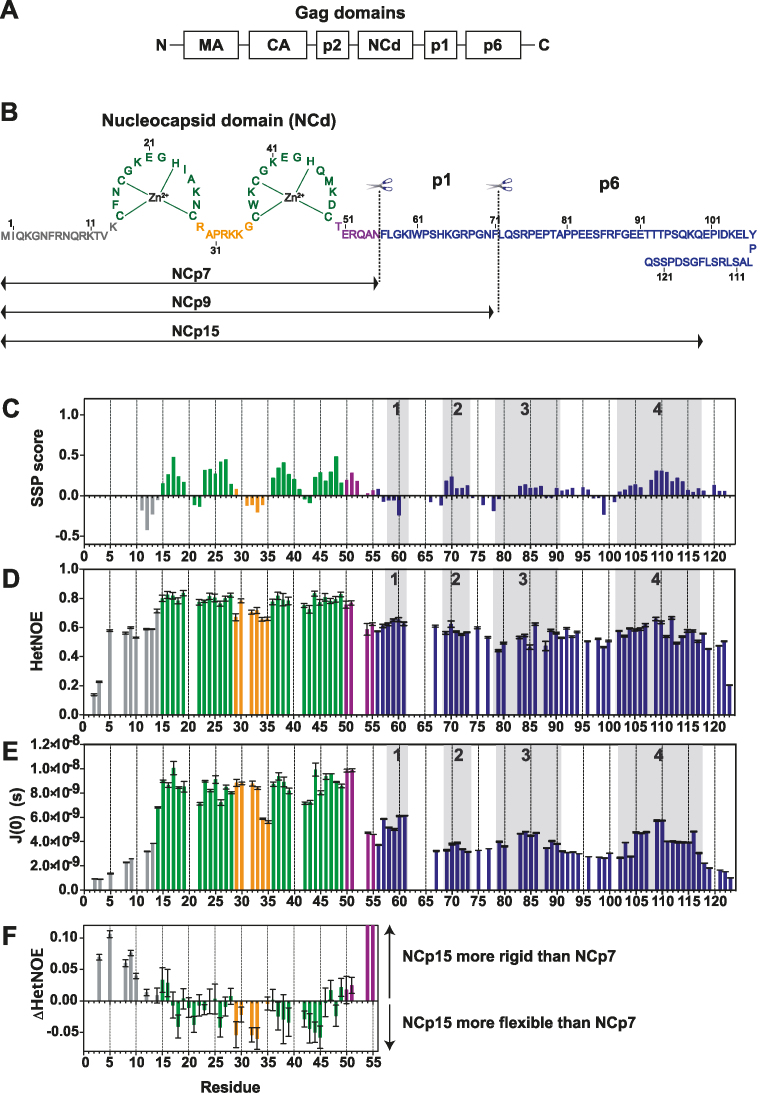
Figure 1.
Structural and dynamic characterization of NCp15 from NMR chemical shifts and 15N relaxation data. (A) Schematic of full-length Gag, (B) Sequence of the HIV-1 C-terminal domain of Gag (NCp15). The first cleavage by the HIV-1 protease first liberates the NCp15 protein, then NCp9 and finally the mature form of NCd called NCp7. The dashed line represents the two sites of protease cleavage present in NCp15. Residues are coloured in grey for residues in the N-terminal part of NCd, green for those in the zinc knuckles and orange for residues in the linker between the two zinc knuckles of NCd and purple for the C-terminal domain of NCd. Due to a limitation in space, the sequence of p6 is not drawn linearly, but this does not represent a fold back of p6 on itself, (C) Secondary structural propensities calculated by the SSP program (58). Cα, C β, CO and Hα were used as input data for the calculation of the SSP score. Positive values indicate the amount of α-helical conformation present along the sequence whereas negative values indicate extended or β-strand conformations. (D) 15N–{1H} NOE (HetNOE) values are indicative of the magnitude of local subnanosecond motions (high values: restricted motions; low values: high-amplitude motions). (E) Spectral densities J(0) extracted by spectral density mapping from 15N relaxation data (T1, T2, HetNOE) of NCp15. J(0) values are indicative of slow overall and segmental tumbling motions present in NCp15. The boxes in grey indicated the four regions of p1–p6 domains showing significant secondary structure propensities. (F) Differences of HetNOE within the NCd between NCp15 and NCp7 to probe the change in restrictions of motions of the backbone of the precursor and mature forms of the NCd.