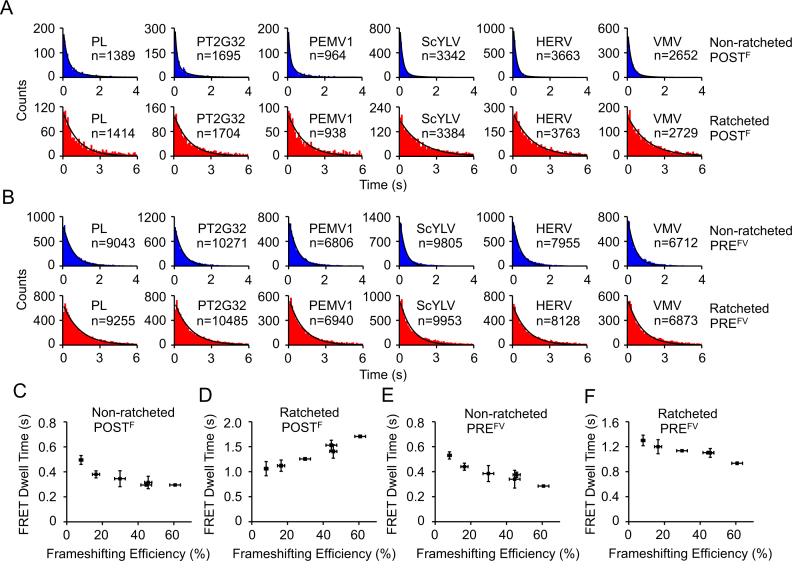
Figure 5.
Conformational dynamics of ribosomal complexes on non-slippery mRNAs with different pseudoknots in the first pseudoknot-unwinding cycle. (A) Dwell time distributions of the non-ratcheted and ratcheted POSTF states. (B) Dwell time distributions of the non-ratcheted and ratcheted PREFV states. (C–F) Plots of dwell times of non-ratcheted POSTF (C), ratcheted POSTF (D), non-ratcheted PREFV (E) and ratcheted PREFV (F) captured by smFRET under equilibrium conditions versus frameshifting efficiency. n is the number of events. Dwell times were extracted by single exponential fitting of dwell time distributions except the non-ratcheted POSTF, whose dwell times were extracted using double exponential decay. Error bars were standard errors.