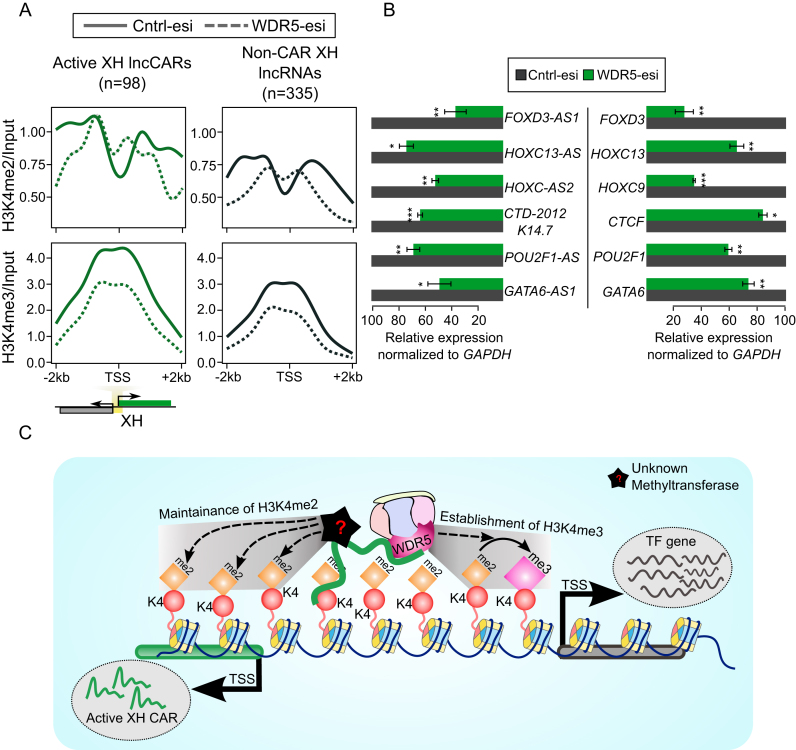
Figure 5.
H3K4me2 and H3K4me3 occupancy upon WDR5 depletion. (A) H3K4me2 and H3K4me3 ChIP-seq signals at the promoter regions (±2 kb) of active XH lncCARs (Green) and non-CAR XH lncRNA (Black) upon WDR5-esi depletion. The signals presented in the plots represent log2ratio between ChIP pull-down and input sample. TSS denotes the transcription start site of active XH lncCARs or non-CAR XH lncRNAs. The solid lines in the plot represents H3K4me2 and H3K4me3 signal in control esiRNA transfected cells and dotted lines represents WDR5 esiRNA transfection. (B) Normalized relative expression of active XH lncCARs (left panel) and their respective partner protein coding genes (right panel) upon removal of WDR5 by esiRNA as compared to that in control esiRNA transfection. Data represent the mean ± SD of two independent biological experiments. * P ≤ 0.05, ** P ≤ 0.01 and *** P ≤ 0.001. (C) Model depicting transcriptional regulation of divergent gene loci by active XH lncCARs. Active XH lncCARs transcribed from divergent transcription units are targeted to specific regions around TSS of partner protein coding gene that are enriched with H3K4me2, through their chromatin interaction property. Promoter targeting of these active XH lncCARs helps in maintaining H3K4me2 in WDR5 independent manner. XH lncCAR-dependent recruitment of WDR5 at the active XH transcription units helps in the conversion of H3K4me2 into H3K4me3, thereby establishing transcriptionally competent chromatin at the divergent transcription units. However, the establishment and maintenance of H3K4me2 levels at these transcription units are probably mediated by a different mechanism.