Figure 1.
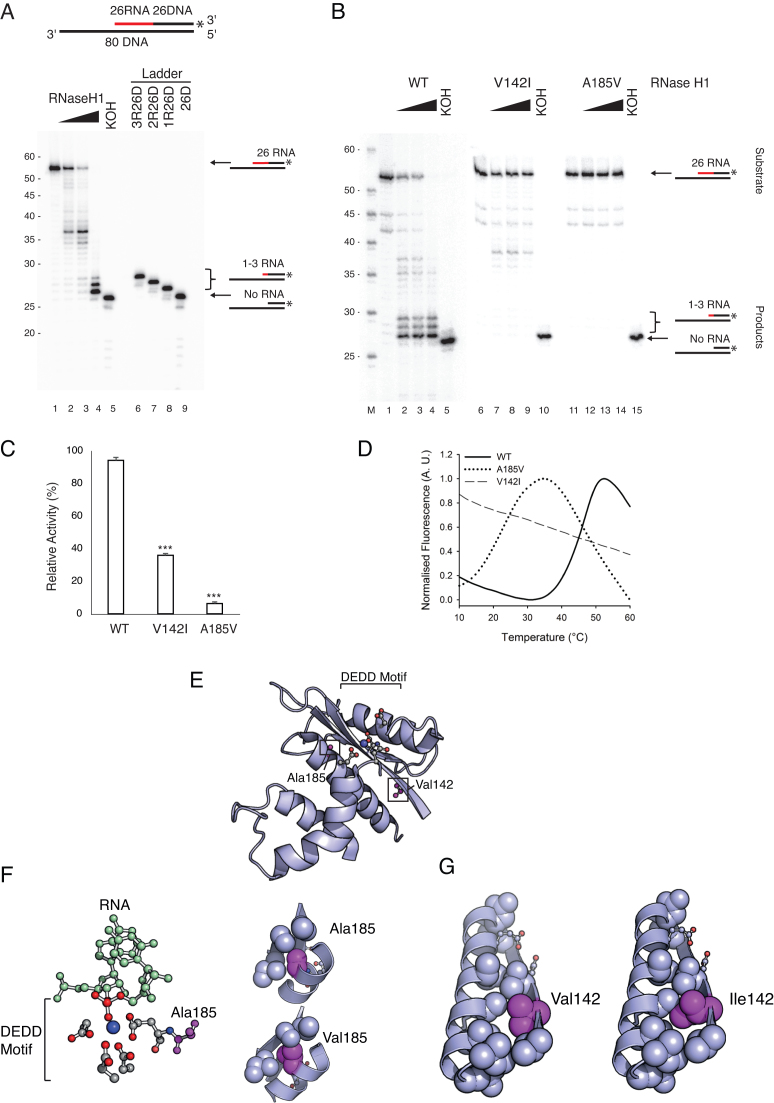
Disease causing mutations in RNase H1 impair primer processing. (A) The cleavage pattern of wild type RNase H1. The 3′-labelled OriL substrate is shown at the top (RNA- red, DNA- black). KOH removes all RNA. 3′-labelled chimeric oligonucleotides were used as a size marker. Wild type RNase H1 efficiently cleaved the RNA near the RNA-DNA junction, but did not cleave the last 1–3 ribonucleotides. (B) The cleavage patterns of wild type and mutant RNase H1. The V142I mutant had decreased activity (∼35-40 nt products); the A185V mutant was inactive. The size marker (M) is labelled at the 5′ end. (C) The endonuclease activity in B was quantified from phosphorimager obtained images: the amount of non-hydrolysed substrate was measured against the negative control input bands (lanes 1, 6 and 11). Mean values ± s.e.m., n = 3, P≤ 1 × 10−3 (Student's unpaired t-test). (D) Thermofluor stability assay shows that the V142I and A185V mutations severely destabilise RNase H1. A185V shifted the Tm by 23.36°C; V142I had severe folding defects, with a near-unfolded state under native conditions. (E) The potential structural/functional consequences of the V142I and A185V mutations were assessed by looking at the crystal structure of wild type RNase H1. Catalytically important residues shown in magenta alongside Ala185 and Val142 (PDB ID: 2QK9). Both residues are located near the active site. (F) Ala185 is adjacent to the catalytically essential Glu186, which coordinates a catalytic Mg2+ ion and forms part of a hydrophobic pocket that mediates the stabilising interactions in the active site region. The A185V mutation causes a steric clash in this pocket. (G) Val142 stabilises the hydrophobic interface between helix α1 and sheet β1. This interface is critical for proper alignment of Asp145 and Glu186 in catalysis which is likely disturbed in the V142I variant.