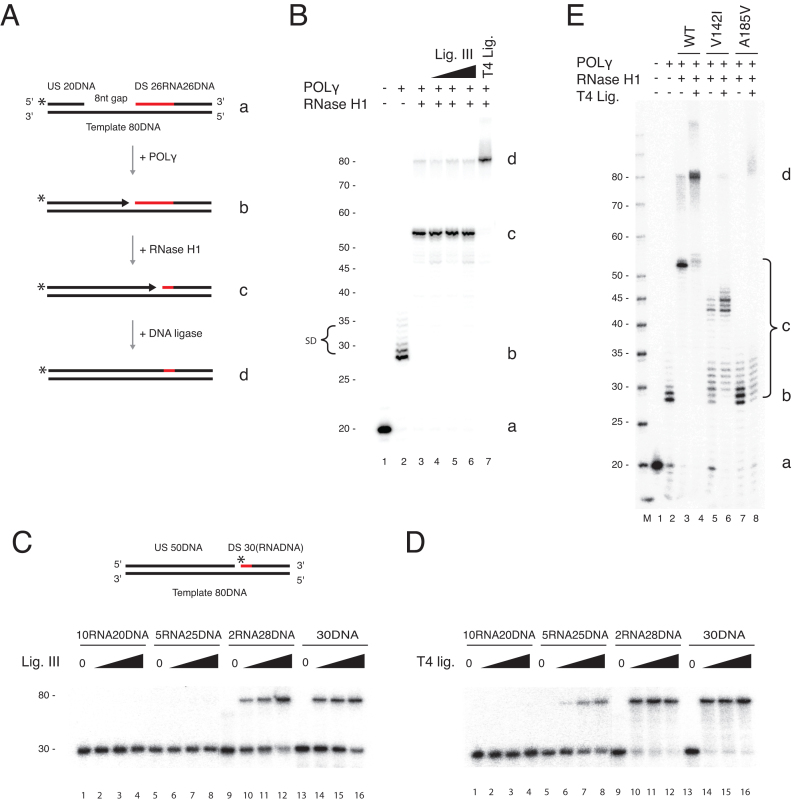
Figure 3.
RNase H1 processing coupled to POLγ dependent DNA synthesis does not produce ligatable nicks. (A) Schematic of the coupled nuclease gap-filling ligation assay performed on a gapped OriL substrate (a). The upstream oligonucleotide was radioactively labelled at the 5′-end. The possible products are illustrated (b–d). (–) Coupled nuclease gap-filling ligation assay as shown in A. POLγ filled the gap (lane 2, marked b) and had limited strand displacement activity (SD). Note though that POLγ completely displaces the downstream oligonucleotide in a small fraction of templates (80 nt band, lanes 3–6,), RNase H1 cleaved the RNA in the substrate (lane 3), enabling further gap-filling (marked c). Only very low levels of ligated products were formed in the presence of 80–320 fmol DNA ligase III (lanes 4–6). A prominent 80 nt ligated product was formed with T4 DNA ligase (lane 7, marked d). The letters a-d correspond to the illustrations in panel A. (C) Ligation assay on a nicked substrate containing RNA tracts of varying length downstream of the nick in the presence of 80–320 fmol DNA ligase III. DNA ligase III discriminates against nicked substrates that contain increasing stretches of ribonucleotides. Two, but not five or more ribonucleotides, can be ligated. (D) As in C, except performed with T4 ligase (1–8 U). T4 ligase can ligate five but not 10 ribonucleotides. (E) T4 ligase-mediated ligation is abolished in the presence of the mutant RNase H1 proteins. The letters a-d correspond to the illustrations in panel A.