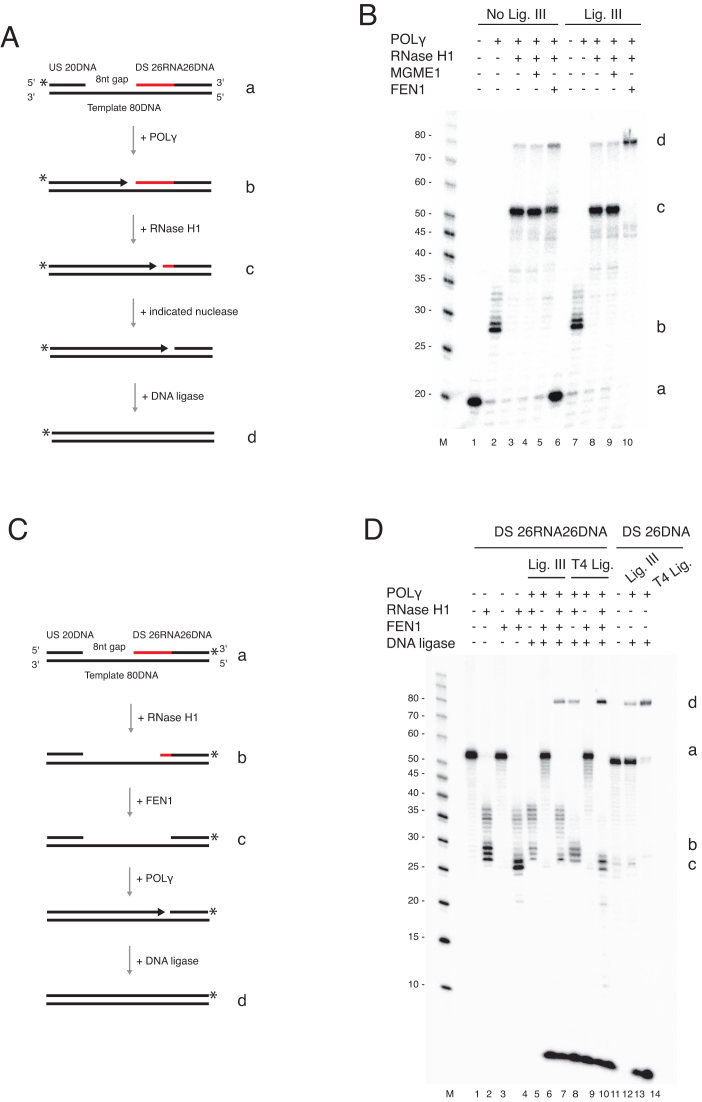
Figure 4.
FEN1 completes primer removal and L-strand maturation. (A) Schematic of template used in panel B. The possible products are illustrated (b–d). (B) Coupled nuclease gap-filling ligation assay was performed as in Figure 3 but in the presence of the indicated nucleases. MGME1 (250 fmol) was added in lanes 4 and 9; FEN1 (35 fmol) was added in lanes 5 and 10. The samples in lanes 6–10 contained 300 fmol DNA ligase III. Conversion of the nicked product (c) to a ligated 80 nt product (d) was stimulated when FEN1 was added together with RNase H1. (C) Schematic of template used in panel D. The downstream chimeric oligonucleotide was labelled at the 3′-end. The possible products are illustrated (b–d). (D) Coupled nuclease gap-filling ligation assay was performed in the presence of RNase H1 alone or together with FEN1 (35 fmol) as indicated. Lanes 2–4 contained only RNase H1 with or without FEN1 to monitor nuclease activity. RNase H1 cleaved the downstream oligonucleotide leaving 1–3 unprocessed ribonucleotides. FEN1 cleaved the remaining ribonucleotides, resulting in a shorter product. POLγ and DNA ligase (ligase III or T4 DNA ligase) were added in lanes 5–10. Ligase III showed ligation only when both RNase H1 and FEN1 were added. However, T4 DNA ligase could ligate without FEN1. Lanes 12–14 are ligation controls using a 3′-end labelled DNA-only oligonucleotide with a 5′-end phosphate. Note that the short band (<10 nt) is the result of POLγ idling at the 3′-end of the template. During the idling process the 3′-5′ exonuclease activity of POLγ cleaves off the 3′-end label.