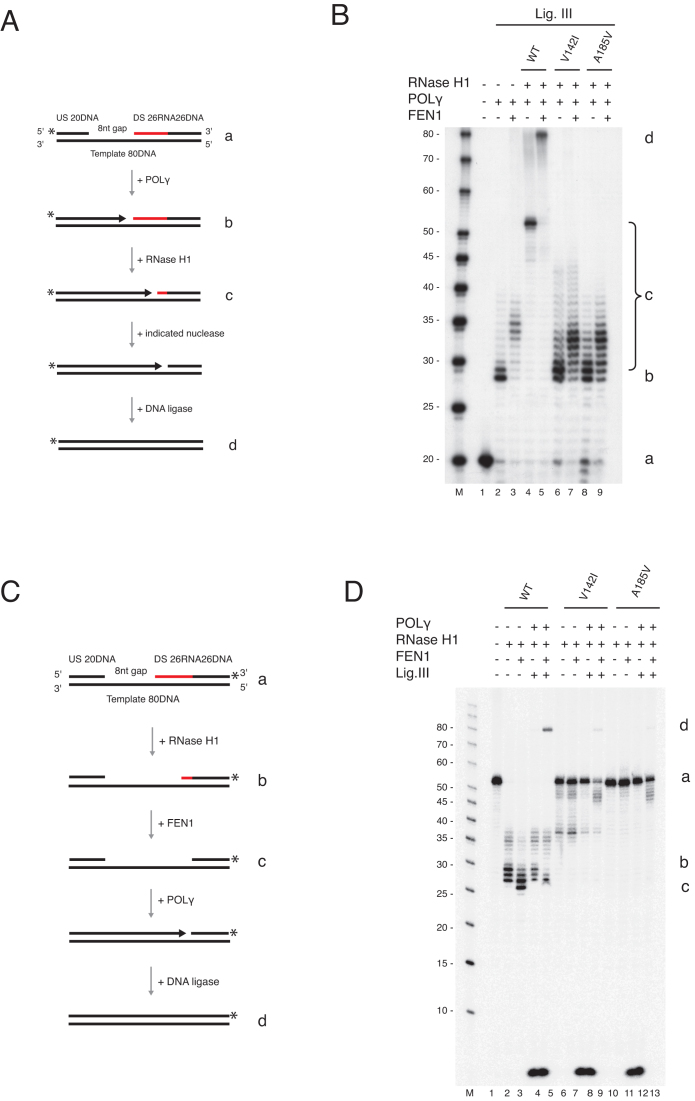
Figure 5.
FEN1 is not able to substitute for RNase H1 during primer removal and L-strand maturation. (A) Schematic of template (a) used in panel B and C. The possible products are illustrated (b–d). The upstream oligonucleotide was radioactively labelled at the 5′-end. (B) Nick translation assay with POLγ and either FEN1 (30, 60, and 120 fmol) or MGME1 (50, 150 and 300 fmol). Partial removal of ribonucleotides was achieved only when FEN1 was used. Lanes 6 and 12 are controls containing only RNase H1. (C) Coupled nuclease gap-filling ligation assay showing that a ligated 80 nt product was only produced when FEN1 was present with the wild type RNase H1 protein. No ligation was seen with FEN1 alone, or with the V142I or A185V mutants. (D) Schematic of template used in panel E. The downstream chimeric oligonucleotide was labelled at the 3′-end. The possible products are illustrated (b–d). (E) Coupled nuclease gap-filling ligation assay was performed as in Figure 4D but only ligase III was used. Wild type RNase H1 was added in lanes 2–5. Lanes 6–9 and 10–13 contained V142I and A185V respectively. The ligation was only observed when wild type and RNase H1 together with POLγ and ligase III was used. Negligible ligation was observed when V142I mutant was used in the presence of FEN1. Note that the short band (<10 nt) is the result of POLγ idling at the 3′-end of the template. During the idling process the 3′-5′ exonuclease activity of POLγ cleaves off the 3′-end label.