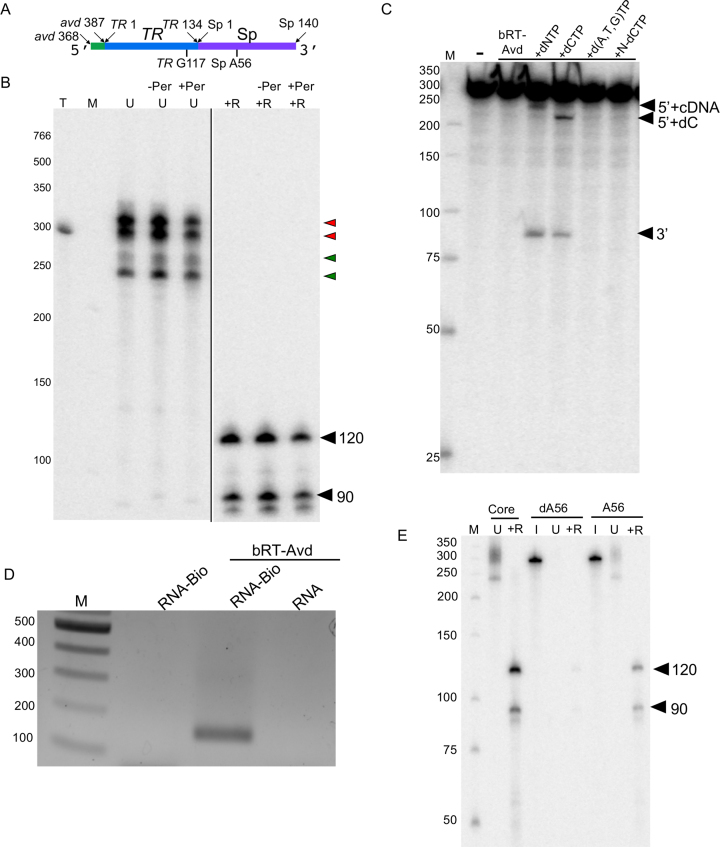
Figure 5.
Core DGR RNA. (A) Schematic of core DGR RNA. (B) Radiolabeled products resulting from bRT-Avd activity for 2 h with the core DGR RNA as template. Prior to the reverse transcription reaction, the RNA template was untreated (-Per) or treated with periodate (+Per). Products from the reaction were untreated (U) or treated with RNase (+R), and resolved by 6% denaturing PAGE. Lane T corresponds to internally-labeled core DGR RNA as a marker for the size of the template. Red arrowheads indicate radiolabeled product bands that migrate at the same position or slower than the core DGR RNA, and green arrowheads ones that migrate faster. The positions of the 120 and 90 nt cDNA bands are indicated. The two panels are from the same gel, with the black line indicating that intermediate lanes were removed. (C) Internally-labeled core DGR RNA was not incubated (–), or incubated with bRT-Avd alone or bRT-Avd with 100 μM standard dNTPs (+dNTP), 100 μM dCTP (+CTP), 100 μM dNTPs excluding dCTP (+d(A,T,G)TP), or 100 μM nonhydrolyzeable analog of dCTP (+N-dCTP) for 2 h. Incubation products were resolved by denaturing PAGE. The band corresponding to the 5′ fragment of the cleaved core RNA containing either a deoxycytidine alone (5′+dC) or cDNA (5′+cDNA), and the band corresponding to the 3′ fragment of the RNA are indicated. (D) The core DGR RNA was biotinylated at its 3′ end (RNA-Bio), and either reacted with no protein or used as a template for reverse transcription with bRT-Avd. The core DGR RNA in its unbiotinylated form (RNA) was also used as a template for reverse transcription with bRT-Avd. Samples were then purified using streptavidin beads, and the presence of TR-cDNA in the purified samples was assessed by PCR. Products from the PCR reaction were resolved on an agarose gel. (E) Radiolabeled products resulting from bRT-Avd activity for 12 h with core, hybrid core dA56, or hybrid core A56 DGR RNA as template. Products were untreated (U) or treated with RNase (+R), and resolved by denaturing PAGE. Separate samples of core dA56 and A56 were 5′ 32P-labeled for visualization of inputs (I). The positions of the 120 and 90 nt cDNAs are indicated.