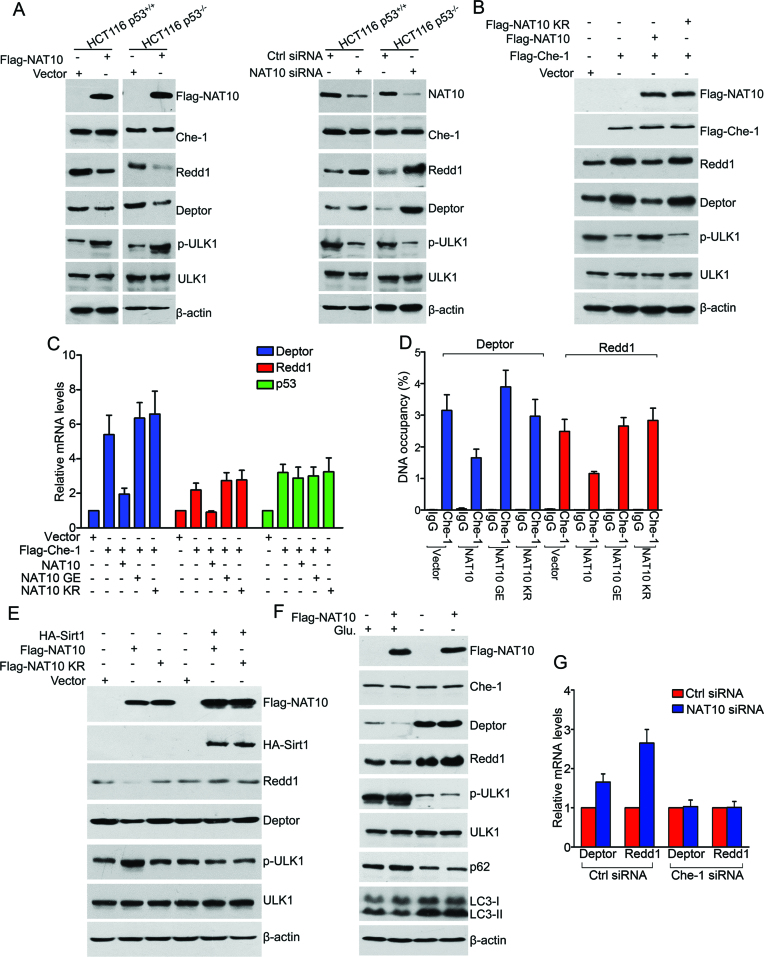
Figure 4.
NAT10 inhibits the Che-1-dependent transcriptional activation of Deptor and Redd1 under energy-rich conditions. (A) HCT116 cells were transfected with the indicated siRNAs or plasmids. Western blotting was performed to evaluate the indicated proteins. (B) HCT116 cells were transfected with the indicated plasmids. After 24 h, cell lysates were prepared and subjected to Western blot analysis for evaluation of the indicated proteins. (C) HCT116 cells were transfected with the indicated plasmids. After 24 h, total RNA was extracted and subjected to RT-qPCR for evaluation of the mRNA levels of the indicated genes. Error bars indicate the SEM (n = 3). (D) HCT116 cells were transfected with the indicated plasmids. The promoter occupancy of Deptor or Redd1 by Che-1 was analyzed by a ChIP assay. Error bars indicate the SEM (n = 3). (E) HCT116 cells were transfected with the indicated plasmids. After 24 h, the cells were harvested, and western blotting was performed to evaluate the indicated proteins. (F) U2OS cells were transfected with the indicated plasmids and cultured in medium with or without glucose for 36 h. The cells were harvested, and Western blotting was performed to evaluate the indicated proteins. (G) HCT116 cells were transfected with the indicated siRNAs. After 72 h, total RNA was extracted and subjected to RT-qPCR for evaluation of the Deptor and Redd1 mRNA levels. Error bars indicate the SEM (n = 3). See also Supplementary Figure S3.