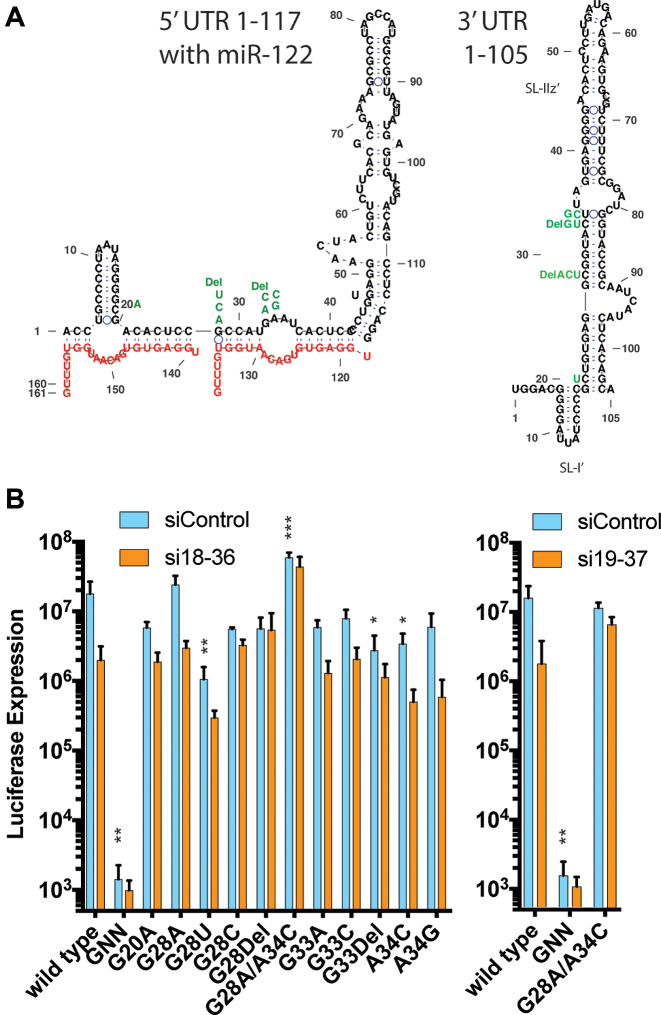
Figure 5.
Sequences of 5′UTR and transient replication assays of full length viral RNA having mutations to evolutionarily variable sites in siRNA target regions. (A) Sequence mutations are shown aligned with the canonical 5′ UTR in the presence of miR-122, and the complementary mutations in the structure of the negative strand 3′ terminus. (B) Transient HCV replication assay analysis of the mutant virus RNA in wild-type Huh-7.5 cells. Luciferase expression was measured as a proxy for genome amplification in cells transfected with full length HCV genomic RNA carrying a luciferase reporter gene and the indicated mutation. Data are the average of three independent transfections and error bars represent the standard deviation. The differences in replication fitness of the mutant viruses versus wild-type (miControl) at each time point was analyzed by one-way ANOVA and P-values are indicated as follows * 0.05 to 0.005, ** 0.005 to 0.0001 and ***<0.0001. No notation indicates no significant difference.