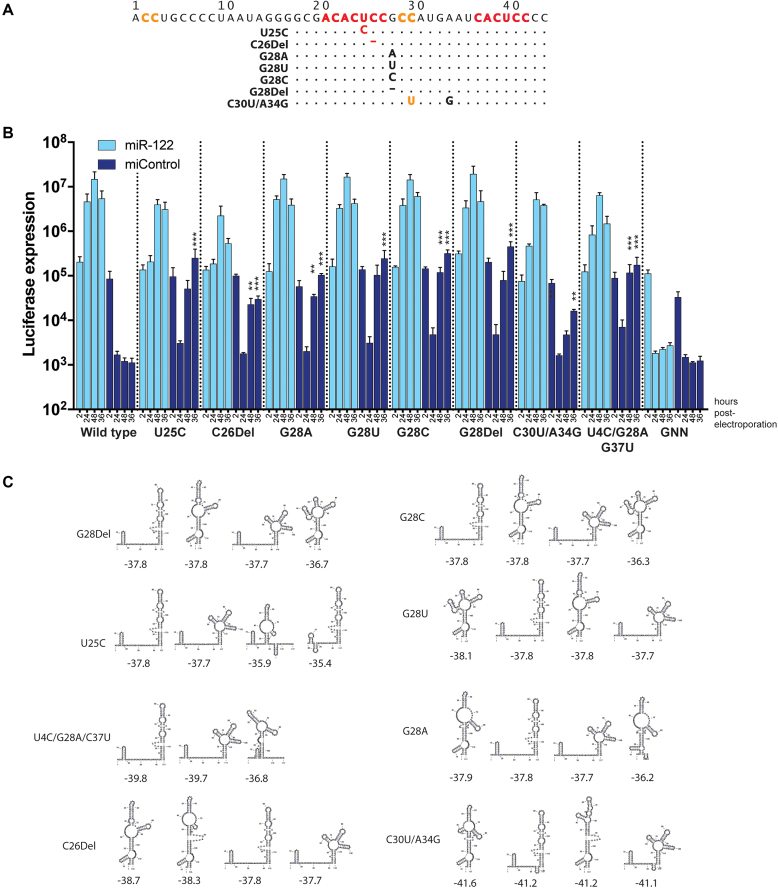
Figure 7.
Sequence and replication analysis of viruses capable of miR-122-independent replication. (A) The 5′ UTR sequences of viruses capable of miR-122-independent replication. (B) Time course analyses of replication of these mutants in miR-122 knockout cells following co-electroporation with the indicated miRNAs (miR-122 or miControl). Data are the average of three independent transfections or electroporations and error bars represent the standard deviation. Significant differences in miR-122-independent replication of the mutants versus wild-type RNAs were calculated using Student’s t-tests for (B) and P-values are indicated as follows * 0.05 to 0.005, ** 0.005 to 0.001, *** 0.001 to 0.005, and, ****<0.0005, no notation indicates no significant difference. (C) RNA structure predictions for the 5′ UTR sequences in the absence of miR-122. The four lowest free energy structures are shown.