Abstract
The different steps of gene expression are intimately linked to coordinate and regulate this complex process. During transcription, numerous RNA-binding proteins are already loaded onto the nascent mRNA and package the mRNA into a messenger ribonucleoprotein particle (mRNP). These RNA-binding proteins are often also involved in other steps of gene expression than mRNA packaging. For example, TREX functions in transcription, mRNP packaging and nuclear mRNA export. Previously, we showed that the Prp19 splicing complex (Prp19C) is needed for efficient transcription as well as TREX occupancy at transcribed genes. Here, we show that the splicing factor Mud2 interacts with Prp19C and is needed for Prp19C occupancy at transcribed genes in Saccharomyces cerevisiae. Interestingly, Mud2 is not only recruited to intron-containing but also to intronless genes indicating a role in transcription. Indeed, we show for the first time that Mud2 functions in transcription. Furthermore, these functions of Mud2 are likely evolutionarily conserved as Mud2 is also recruited to an intronless gene and interacts with Prp19C in Drosophila melanogaster. Taken together, we classify Mud2 as a novel transcription factor that is necessary for the recruitment of mRNA-binding proteins to the transcription machinery. Thus, Mud2 is a multifunctional protein important for transcription, splicing and most likely also mRNP packaging.
INTRODUCTION
Gene expression is a fundamental cellular process, without which life is impossible. It consists of multiple steps starting with transcription, i.e. the synthesis of the mRNA by RNA polymerase II (RNAPII). Already co-transcriptionally the mRNA is processed: At the 5′ end a cap structure is added, introns are spliced out and—after cleavage at the 3′ end—a poly(A) tail is added. In addition, the mRNA is packaged into a messenger ribonucleoprotein particle (mRNP) by nuclear mRNA binding proteins (1–3). Only fully processed and packaged mRNPs are exported from the nucleus to the cytoplasm by the conserved mRNA exporter called Mex67-Mtr2 in Saccharomyces cerevisiae and TAP-p15/NXF1-NXT1 in metazoans (4–7). Interestingly, different steps of gene expression are intimately linked for regulation and quality control. Importantly, the composition of the mRNP sometimes determines the fate of the mRNA at later, e.g. also cytoplasmic, stages.
Several nuclear mRNA-binding proteins already bind co-transcriptionally to the mRNA such as the SR-like proteins Nab2 and Npl3 or Tho1 in S. cerevisiae and thus constitute the nuclear mRNP [(1) and references therein]. The recruitment of proteins involved in mRNA processing and mRNP packaging is coordinated with the different phases of transcription by the C-terminal domain (CTD) of Rpb1, the largest subunit of RNAPII (1,8,9). The CTD consists of repeats of the heptapeptide sequence YSPTSPS. The number of repeats differs between organisms: There are, for example, 52 repeats in humans and 26 repeats in S. cerevisiae. In addition to other modifications, the CTD becomes differentially phosphorylated with the progression of the transcription cycle. During transcription initiation, mainly serine 5 (S5) and serine 7 (S7) are phosphorylated, whereas during elongation, tyrosine 1 (Y1) and serine 2 (S2) phosphorylation is high, which decrease at the poly(A) site and the termination site, respectively (10–12). Consequently, factors involved in splicing and mRNP packaging were shown to bind to the S2 phosphorylated CTD (also see below).
One of the protein complexes important for nuclear mRNP formation is the TREX complex. In S. cerevisiae, TREX is a heterononameric complex consisting of the THO subcomplex (composed of Tho2, Hpr1, Mft1, Thp2 and Tex1), the SR-like proteins Gbp2 and Hrb1, the DEAD-box helicase Sub2 and Yra1 (1,4–6,13–16). TREX couples transcription to nuclear mRNA export as it is important for transcription elongation and also recruits the mRNA exporter Mex67-Mtr2 to the mRNA (13,17–21). In addition to TREX, the SR-like proteins Nab2 and Npl3 recruit Mex67-Mtr2 to the mRNA (1,22). Furthermore, TREX is involved in 3′ end processing and transcription-coupled DNA repair and prevents hyperrecombination (23–25). TREX interacts with the transcription machinery via the binding of its subunit Yra1 to Pcf11, a component of the 3′ end processing machinery, and the S2 phosphorylated CTD (26,27). We previously showed that TREX is recruited to the transcription machinery by the direct interaction of its subcomplex THO with the S2 phosphorylated CTD (28). The association of TREX with the transcription machinery is maintained by the Prp19 complex (Prp19C) (29,30). Prp19C was first shown to function in splicing, which is carried out by the spliceosome composed of small nuclear ribonucleoprotein particles (snRNPs) and additional non-snRNP proteins. Prp19C was also named nineteen complex (NTC) and is a non-snRNP protein complex (31,32). Prp19C’s function in splicing and TREX occupancy at the transcribed gene are probably independent of each other as a mutation in the Prp19C component Syf1 causes a decreased occupancy of TREX but does not display a splicing defect (29). Probably due to its function in TREX occupancy, Prp19C is necessary for efficient transcription (29). Taken together, Prp19C and TREX function in transcription elongation as well as nuclear mRNP formation.
As expected from their fundamental functions in gene expression, both TREX and Prp19C are conserved. The human homologs of the THO subunits Hpr1, Tho2 and Tex1 are named THOC1, 2 and 3, respectively. Human THO has three additional subunits THOC 5, 6 and 7 that have no orthologues in budding yeast. Furthermore, the human homologs of the TREX subunits Sub2 and Yra1 are named UAP56 and ALYREF/THOC4. Human TREX functions in transcription and mRNA processing and export (4,15). In addition, TREX is also important for embryogenesis, organogenesis as well as cellular differentiation and implicated in several diseases (15,33). The subunit composition of the TREX complex in Drosophila is similar to the human one (34). Drosophila TREX consists of the core THO subunits as well as THOC5, THOC6, and THOC7. UAP56 and ALYREF are less stably associated with TREX than their yeast homologs Sub2 and Yra1, respectively. Interestingly, nuclear export of the vast majority of mRNAs is independent of TREX but export of heat-shock mRNAs depends on THO function under stress (34). The Prp19 complex is also conserved from yeast to humans (35). The human PRP19/CDC5L complex is a large protein complex including hPrp19, CDC5L (Cef1 in S. cerevisiae), PRL1 (Prp46), SPF27 (Snt309) and several other subunits that are not present in yeast Prp19C (36). In addition to PRP19/CDC5L two other protein complexes containing Prp19 and homologs of some yeast Prp19C subunits have been isolated in humans: the PRP19-associated complex and the XAB2 complex (36,37). In addition to pre-mRNA splicing, human Prp19C subunits are involved in transcription and transcription-coupled repair (37). In Drosophila, two Prp19 complexes exist that are homologous to the human PRP19/CDC5L and PRP19-associated complexes (38). Drosophila Prp19C is associated with the spliceosome and is required for splicing (39). Furthermore, Prp19C is essential for Drosophila development and mutation of its subunits causes different developmental defects. For example, mutation of the subunit Fandango, the homolog of yeast Syf1, disrupts blastoderm cellularization (38), while mutation of CRN1, the homolog of yeast Syf3, causes early embryonic lethality (40).
Despite many insights gained during the past couple of years, the process of nuclear mRNP formation is still poorly understood. Specifically, our knowledge about the multiple interactions between the proteins and protein complexes necessary to recruit mRNA binding proteins to the mRNA in order to form an mRNP remains fragmentary. Interestingly, U2AF65, the potential human homolog of the yeast protein Mud2, recruits Prp19C to the nascent mRNA by binding directly to the phosphorylated CTD as well as Prp19C and thus enhances splicing in in vitro splicing assays with an SRSF1-CTD fusion protein that recruits the CTD to the splicing substrate (41). During splicing Mud2 associates with the pre-mRNA early during spliceosome assembly, recognizes—together with its binding partner the branch-point binding protein BBP/Msl5—the branch point (BP) sequence and bridges between the BP and the U1 snRNP at the 5′ splice site (42–44). Here, we show that Mud2 also interacts with Prp19C in S. cerevisiae. Furthermore, Mud2 is necessary to recruit Prp19C and TREX to the transcribed gene in a S2 phosphorylation dependent manner. Importantly, Mud2 is also recruited to intronless genes and required for full transcriptional activity in vivo and in vitro. Taken together, we uncover a novel role for Mud2 in gene expression: Mud2 functions in transcription most likely via recruitment of Prp19C and TREX.
MATERIALS AND METHODS
Strains, plasmids and primers
Yeast strains, plasmids and primers are listed in Supplementary Tables S1, S2 and S3, respectively.
Generation of antibodies
Antibodies against Drosophila melanogaster Prp19 (fragment encompassing amino acids 1–232), Fandango (fragment encompassing amino acids 542–750) and U2AF50 (fragment encompassing amino acids 1–415) were raised by immunizing rabbits with the corresponding His6-tagged protein fragments. These antibodies were affinity-purified using the respective antigen coupled to cyanuric chloride-activated sepharose (Supplementary Figure S4A–C).
Chromatin immunoprecipitation experiments
Chromatin immunoprecipitation (ChIP) experiments in S. cerevisiae were performed according to (45) with some modifications. Briefly, 100 ml yeast culture in mid-log phase were crosslinked. The lysate was sonicated using a Bioruptor UCD-200 (Diagenode) for 3 × 15 min (30 s ON/30 s OFF) at ‘HIGH’ power setting with intermittent cooling resulting in chromatin fragments of 200–250 bp. TAP-tagged proteins were immunoprecipitated with IgG-coupled Dynabeads (tosylactivated M280, Thermo Scientific) for 2.5 h at 20°C. For ChIPs of RNAPII and S2P, the monoclonal antibody 8WG16 (Biolegend) or 3E10 (46), respectively, was added for 1.5 h at 20°C followed by 1 h incubation with Protein G Dynabeads. For RNAPII ChIPs in the S2A mutant a polyclonal antibody directed against the N-terminal domain of Rpb1 (yN-18, Santa Cruz Biotechnology) was added for 1.5 h at 20°C followed by 1 h incubation with Protein G dynabeads. A non-transcribed region (NTR1, 174131–174200 on chr. V) served as negative control. The occupancy of each protein was calculated as its enrichment at the respective gene relative to NTR1.
ChIP experiments with Drosophila Schneider 2 (S2) cells were performed according to (47). The DNA was crosslinked (1% formaldehyde, 15 min) and sheared to a size of ∼300 base pairs (bp). Approximately 3 × 106 cells and 10 μg of each antibody (RNAPII: Abcam) were used for one experiment. The recovered DNA was analyzed by qPCR with a Light Cycler 96 (Roche) at six positions along the hsp70 gene: −168, −43, 648, 974, 1698 and 1940 bp. Sequences of the primers used are available on request. Each ChIP was repeated at least five times. As negative control, ChIP experiments with preimmune IgG were performed. To induce expression of the hsp70 gene S2 cells were exposed to heat shock for 20 min at 37°C.
Protein purification
TAP-tagged proteins were purified from S. cerevisiae using the tandem affinity purification (TAP) technique as described (13,29,48). To determine proteins co-purifying with Mud2 a strain expressing flag-TEV-protA tagged Mud2 was used (49). To assess the RNA-dependency of the interaction, 100 μg/mL of RNase A (Sigma) were added to the lysates during the incubation with IgG-coupled sepharose beads. For the CTD binding assay Mud2, Pcf11 and the Rix1 complex were purified until the TEV eluate using IgG-coupled tosylactivated magnetic beads.
For the add-back transcription assays, Mud2 was either purified from S. cerevisiae by TAP until the TEV eluate using the MUD2-TAP strain as described above, except that the IgG-coupled sepharose beads were washed with TAP buffer containing 250 mM NaCl after binding. To purify recombinant Mud2, Mud2-His6 was expressed in E. coli Rosetta 2 (DE3), and cells were lysed in lysis buffer (50 mM KPi pH 8.0, 200 mM NaCl, 0.1% Triton X-100, 10 mM ß-mercaptoethanol, protease inhibitors) using sonication. The lysate was incubated for 1 h at 4°C with Ni-NTA agarose beads (Qiagen) and applied to an ‘Econo-Column’ glass chromatography column (Biorad) equilibrated with buffer. The column was washed 4 times with 1 ml wash buffer (lysis buffer without Triton X-100; containing 20 mM imidazole). Mud2 fractions were eluted from the column using wash buffer with 150 mM imidazole. Peak fractions were collected, aliquots frozen in liquid nitrogen and stored at −80°C.
For the immunoprecipitation experiments with Drosophila proteins, Drosophila Schneider line 2 (S2) cells were maintained at 25°C in Schneider's Insect Medium (Sigma) containing 10% fetal bovine serum (HyClone, USA). To extract proteins, S2 cells were lysed in 10 mM HEPES, pH 7.9, 5 mM MgCl2, 0.5% NP-40, 0.45 M NaCl, 1 mM DTT and complete protease inhibitor cocktail (Roche). Immunoprecipitation was performed as described previously (50). Treatment of the extract with DNase I (USB, 0.6 U/ml) and RNase (Stratagene, 10 U/ml) was performed at 37°C for 20 min. In addition to the antibodies generated for this study the following antibodies were used: TAF3 antibody (51), RNAPII antibody (52), and S2P antibody (Abcam, phospho S2, Ab5095).
CTD-peptide pulldown assay
Pulldown assays were performed as described in (28). Briefly, TEV eluates from wild-type, MUD2-TAP, PCF11-TAP and RIX1-TAP strains were incubated with non-phosphorylated or S2 phosphorylated biotinylated CTD peptides (PSL, Heidelberg) bound to Streptavidin-coupled magnetic beads (M280, Invitrogen). After the binding reaction, beads were washed four times. The peptide-bound proteins were eluted by boiling the beads (98°C) for 3 min in 20 μl of 2× SDS sample buffer. Equal amounts of each sample were loaded for Western blotting. Bound and unbound proteins were detected with an anti-CBP antibody (Open Biosystems, CAB 1001).
Quantitative Western blots
To quantify the total amount of a protein, cells were grown to mid-log phase and lysed by the NaOH method and proteins precipitated with TCA (53). Equal amounts of total protein were separated by SDS-PAGE, and proteins were detected with the corresponding primary antibody, a horse radish peroxidase-coupled secondary antibody and CheLuminate-HRP PicoDetect ECL solution (Applichem). Bands were imaged using a ChemoCam Imager (Intas) and quantified using the LabImage 1D Gel Analysis Software (Intas).
In vivo transcription assay
Cells were grown in media containing raffinose to an OD600 of 0.8, and transcription from the GAL10 promoter was induced by addition of 2% galactose for the indicated length of time. Total cellular RNA was isolated from 25 ml cultures by standard hot phenol extraction. The amount of GAL10 mRNA, ACT1 mRNA and the RNAPIII transcript SCR1 was measured by a primer extension assay using 5′-Cy5-labeled reverse primers specific for GAL10, ACT1 and SCR1 according to (29) with 10 μg, 5 μg and 5 μg of total RNA for GAL10, ACT1 and SCR1 reverse transcription reactions, respectively. The cDNA was separated on a 7 M urea 7% polyacrylamide gel and quantified using a Typhoon 9400 and ImageQuant software (GE Healthcare).
In vitro transcription assay and add back
Yeast nuclear extracts were prepared from 6 L cultures essentially as described (54) with some modifications. The resuspension buffer contained 50 mM Tris pH 7.5, 20 mM EDTA, 30 mM DTT. Cells were spheroplasted by adding 3 ml resuspension buffer containing 18 mg Zymolyase-100T (Seikagaku) and protease inhibitor cocktail (as used for TAP). The protein concentration of each nuclear extract was measured by Bradford and equal concentrations were used from the wild-type and mutant cells for the in vitro transcription assays.
In vitro transcription assays were performed with a plasmid template LL279 containing the HIS4 promoter (–428 to +24 respective to the start codon) as described previously (29,54) with few changes. Transcription reactions were performed in 50 μl volumes at 18°C or 23°C for 1 h. Each reaction mixture contained 200 μg yeast nuclear extract, 200 ng template plasmid, 0.1 M DTT, 64 mg/ml phosphocreatine, transcription buffer (10 mM HEPES pH 7.6, 50 mM potassium acetate, 0.5 mM EDTA, 2.5 mM magnesium acetate), 0.2 μg creatine phosphokinase, 10 U RiboLock RNase inhibitor (Fermentas), 200 ng recombinant Gcn4 and 100 μM nucleoside triphosphates (NTPs). RNA was isolated with a standard phenol chloroform extraction procedure. Transcripts were analyzed by primer extension assays (54) using a 5′-Cy5-labeled primer. Quantification was performed with a Typhoon 9400 scanner and the ImageQuant software (GE Healthcare).
Add-back experiments were performed by adding either Mud2 purified from S. cerevisiae or recombinant Mud2 purified from Escherichia coli.
Analysis of PAR-CLIP data
RNA-Seq normalized PAR-CLIP data of mRNP biogenesis factors in S. cerevisiae (55) were downloaded from the Gene Expression Omnibus (56) (GSE59676). CLIP sites were intersected with the S. cerevisiae (sacCer3) from Saccharomyces Genome Database (SGD) (57) using Bedtools (v.2.25.0) in a strand specific manner. The occupancy values for each factor were accumulated for the exonic region of intronless transcripts, i.e. the whole transcript, and for intronic regions of intron-containing transcripts. Accumulated occupancy values were further normalized to the intronless and intronic region size, resulting in a normalized occupancy value for both regions (intronless and intronic) for each protein. The following number of intronless transcripts and introns contained CLIP tags for the respective protein: Ist3 (intronless/intron: 3647/214), Luc7 (4942/230), Msl5 (5503/240), Mud1 (4301/219), Mud2 (5563/247), Nam8 (4644/221), Snp1 (3098/175), Hpr1 (5126/168), Nab2 (5411/206), Npl3 (5757/232).
Statistical analysis
All data are presented as average ± standard deviation (error bars) of at least three biologically independent experiments. Asterisks indicate the statistical significance (Student's t-test; *P < 0.05 and **P < 0.01).
RESULTS
Mud2 is recruited to intron-containing and intronless genes in a transcription-dependent manner
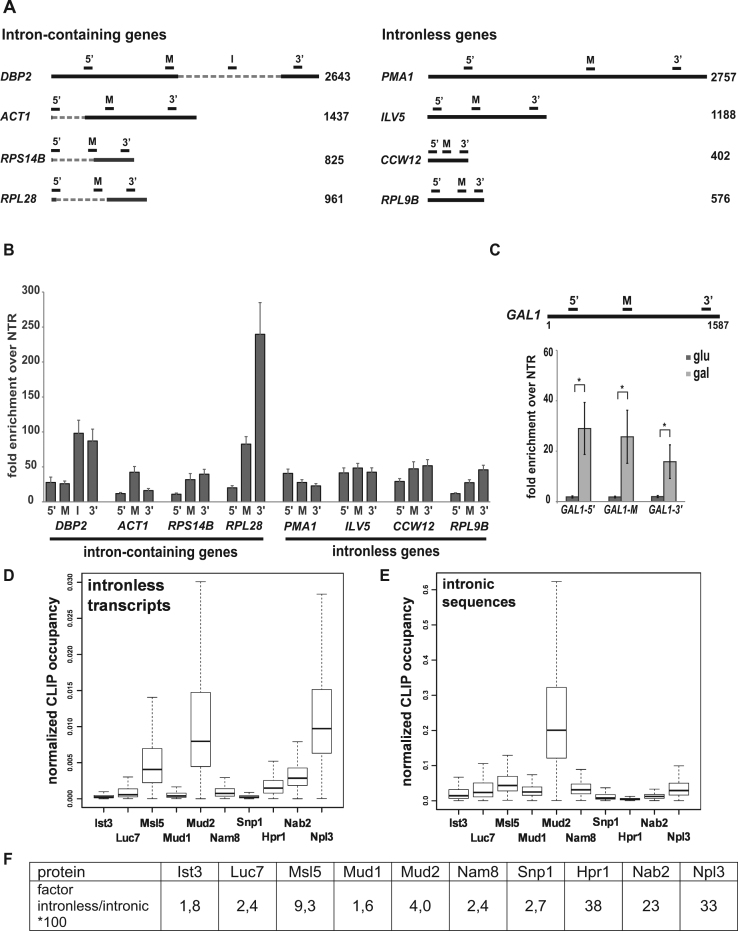
Previously, we showed that the splicing complex Prp19C functions in transcription elongation and ensures occupancy of TREX at transcribed genes (29). In HeLa cells, Prp19C is recruited to the mRNA by U2AF65 in in vitro splicing reactions, in which the CTD is tethered to the splicing substrate by an SRSF1-CTD fusion protein (41). Furthermore, deletion of MUD2, the potential homolog of U2AF65 in S. cerevisiae, suppresses the lethal phenotype caused by deletion of SUB2, a TREX subunit (58). Thus, we wanted to determine whether Mud2 is needed for Prp19C and TREX occupancy at transcribed genes and functions in transcription. If so, Mud2 should be recruited to intron-containing as well as intronless genes, i.e. genes encoding transcripts that are not spliced. We tagged endogenous Mud2 with the TAP-tag and performed chromatin immunoprecipitation (ChIP) experiments at four exemplary intron-containing and four exemplary intronless genes (Figure 1A) to determine the occupancy of Mud2 at transcribed genes. Mud2 is not only present at intron-containing genes but also at intronless genes (Figure 1B). This is consistent with the previous finding that Mud2 is present at the intronless PDR5 gene and binds intronless genes genome-wide (59,60).
Figure 1.
Mud2 is recruited to intronless genes in a transcription-dependent manner. (A) Scheme of the four exemplary intron-containing genes DBP2, ACT1, RPS14B and RPL28 and four exemplary intronless genes PMA1, ILV5, CCW12 and RPL9B. Open reading frames (ORFs) are represented by a solid line and introns by a hatched line. The bars above the genes indicate the position of the primers pairs used for analysis of the ChIP experiments. 5′: 5′ end of the ORF, M: middle, 3′: 3′ end, I: within the intron. (B) Mud2 is not only present at intron-containing but also at intronless genes. The occupancy of Mud2 was analyzed at different positions of four exemplary intron-containing and four exemplary intronless genes by ChIP. The occupancy was calculated as the enrichment of Mud2 at the respective gene relative to its presence at a non-transcribed region (NTR) that served as a negative control (NTR1, 174131–174200 on chromosome V). (C) Recruitment of Mud2 to the gene is transcription-dependent. Occupancy of Mud2 at the GAL1 gene under repressive conditions (cells grown in glucose-containing medium; dark-gray bars) and induced conditions (cells grown in galactose-containing medium; light-gray bars). Primer pairs amplify the 5′-, middle (M) and 3′-region of GAL1 as indicated in the top panel. (D–F) Mud2 binds to intronless transcripts in vivo. Box plots of CLIP hits of Mud2, the splicing factors Ist3, Luc7, Msl5, Mud1, Nam8 and Snp1 as well as the nuclear mRNA-binding proteins Hpr1, Nab2 and Npl3 at intronless transcripts (D) and at the intronic regions of intron-containing transcripts (E). Dashed lines show the lowest and highest values in the dataset. The solid middle line indicates the median value of the data. Boxes mark the lower and upper quartiles, including smallest 25% or 75% of the data values that are smaller/greater than or equal to this characteristic value. The relative binding to intronless transcripts over intronic sequences of intron-containing transcripts of each factor is given in the table (F).
To determine whether Mud2 occupancy at the transcribed gene is dependent on transcription we assessed Mud2 occupancy at the regulatable GAL1 gene (Figure 1C, upper panel). In glucose-containing medium the GAL1 gene is repressed, whereas in galactose-containing medium transcription of GAL1 is induced. Mud2 is not recruited to the GAL1 gene under repressed conditions (Figure 1C, dark-gray bars, glucose). When transcription is induced, Mud2 occupancy at the GAL1 gene significantly increases (Figure 1C, light-gray bars, galactose). Taken together, Mud2 is recruited to intronless genes in a transcription-dependent manner.
As Mud2 is an RNA-binding protein and recruited to intronless genes, we wanted to know, whether Mud2 binds to intronless mRNAs in vivo. PAR-CLIP experiments revealed that Mud2 preferentially binds to unspliced, i.e. not yet spliced, rather than spliced mRNAs in vivo (55). Mud2 binds to the whole intronic sequence with a peak at the pyrimidine/U-rich region 15 nt downstream of the branchpoint (BP) sequence similar to the BP binding protein Msl5 and the U2 snRNP component Ist3, which bind the whole intronic sequence with a peak at the BP and 27 nt downstream of the BP, respectively (55). Thus, Mud2 binds to introns in a manner similar to U2AF65, as U2AF65 binds to the polypyrimidine tract (61). However, this study did not compare the binding intensity of Mud2 to intron-containing versus intronless mRNAs. Thus, we calculated the number of CLIP hits of Mud2 mapping to intronless transcripts versus the intronic regions of intron-containing transcripts normalized to the abundance of each transcript and sequence length (Figure 1D and E). We did not include Mud2 binding to the exonic sequences of intron-containing transcripts in this analysis in order to avoid including spill-over effects from intron binding as binding to exonic sequences and/or recruitment to exonic sequences based on the presence of an intron. We compared the binding of Mud2 to intronless transcripts and to intronic sequences to the binding intensities of all the other splicing factors analyzed in the same study as well as the THO component Hpr1 and the nuclear mRNA-binding proteins Nab2 and Npl3 (Figure 1D and E). To compare the relative binding intensities of all of these factors we calculated the ratio of binding to intronless transcripts over intronic sequences of intron-containing transcripts (Figure 1E). Compared to the other splicing factors (Ist3, Luc7, Nam8, Snp1) Mud2 has the highest ratio of binding to intronless mRNAs with the exception of its binding partner Msl5 (Figure 1D). However, this ratio is even higher for the TREX component Hpr1 and the SR-like proteins Nab2 and Npl3, all of which function in nuclear mRNA packaging (Figure 1E). Within transcripts Mud2 preferentially binds the 5′ region (55). Taken together, Mud2 is recruited to intronless genes and binds to intronless mRNAs in vivo.
Phosphorylation of serine 2 of the CTD is required for Mud2 and Prp19C occupancy in S. cerevisiae
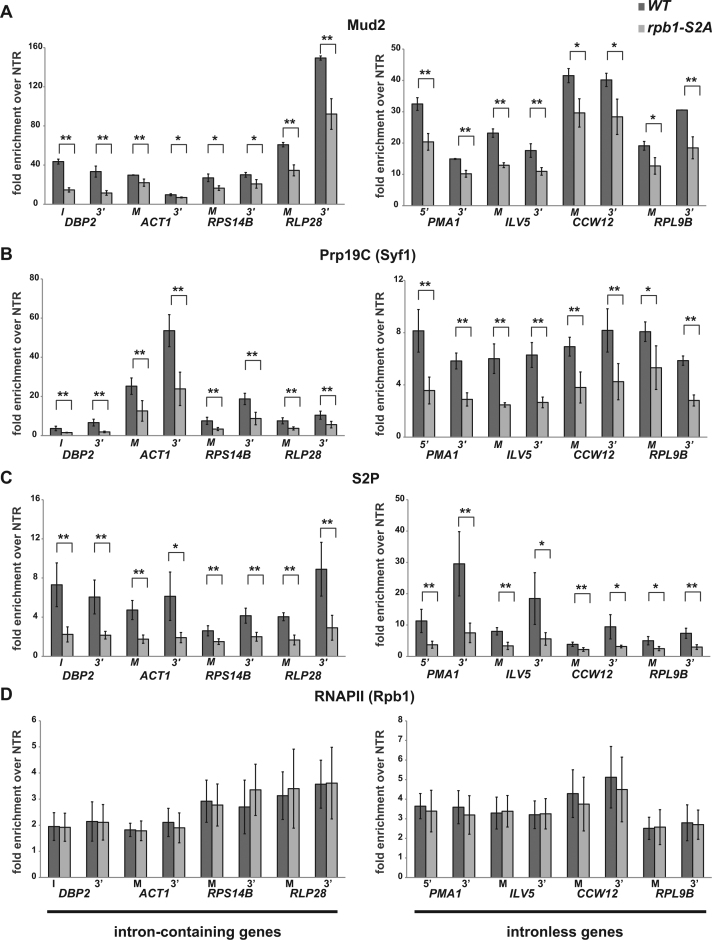
Previously, it was shown in HeLa cells, that tethering of the CTD of the largest subunit of RNA polymerase II (RNAPII) to the splicing substrate enhances splicing by recruitment of U2AF65, which in turn recruits the Prp19 complex (Prp19C) to the mRNA (41). Thus, we wanted to assess whether Mud2 is also recruited to transcribed genes in an S2 phosphorylation dependent manner. We performed ChIP experiments of Mud2 in a serine 2 to alanine (S2A) substitution mutant with nine wild-type and six S2A repeats compared to ‘wild-type’ cells that express Rpb1 with 14 wild-type repeats (62). Mud2 occupancy at these genes is significantly decreased in the S2A mutant compared to wild-type cells (Figure 2A). If Mud2 is needed for recruitment of Prp19C to the gene (also see below), the occupancy of Prp19C should also be decreased in this strain. Indeed, occupancy of the Prp19C subunit Syf1 and thus most likely the whole Prp19 complex is decreased in the S2A mutant (Figure 2B). As expected, the occupancy of RNAPII containing an S2 phosphorylated CTD decreases in the S2A mutant (Figure 2C). In contrast, the decreased occupancy of Mud2 and Prp19C is not due to decreased RNAPII occupancy as the latter is not significantly affected in the S2A mutant (Figure 2D). Thus, serine 2 of the CTD is necessary for recruitment of Mud2 and consequently the Prp19 complex.
Figure 2.
Occupancy of Mud2 and the Prp19C component Syf1 requires S 2 phosphorylation. (A) Mud2 occupancy decreases in the rpb1-S2A mutant. ChIP of Mud2 at two positions of intron-containing and intronless genes according to the scheme shown in Figure 1A and the experiment in Figure 1B. (B–D) The occupancy of the Prp19C subunit Syf1 and of RNAPII with an S2 phosphorylated CTD decreases in the rpb1-S2A mutant, whereas the occupancy of the RNAPII subunit Rpb1 is not affected. ChIP experiments as in (A).
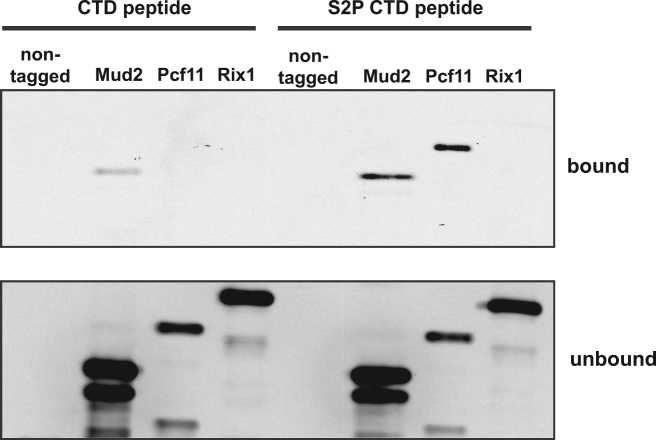
To test, whether Mud2 is recruited to transcribed genes by binding to the S2 phosphorylated CTD, we performed in vitro pulldown experiments. CTD peptides that were either non-phosphorylated or phosphorylated on S2 were immobilized on beads and incubated with Mud2 purified from yeast (Supplementary Figure S1). Pcf11, a 3′ end processing factor that binds to the S2 phosphorylated CTD, served as positive control and the unrelated Rix1 complex, which is required for processing of the 35S pre-rRNA, served as negative control (63,64). Mud2 binds to the S2 phosphorylated CTD and very weakly to the non-phosphorylated CTD (Figure 3, upper panel). This is consistent with the observed binding of U2AF65 to the phosphorylated CTD in vitro (41). However, it was also reported that U2AF65 binding to RNAPII is independent of CTD phosphorylation (65). In summary, Mud2 binds to the S2 phosphorylated CTD. Taken together with the requirement of S2 for Mud2 occupancy in vivo (Figure 2A) S2 phosphorylation is most likely needed for full occupancy of Mud2 and therefore Prp19C at transcribed genes in S. cerevisiae.
Figure 3.
Mud2 binds directly to the S2 phosphorylated CTD. The unrelated Rix1 complex served as negative and Pcf11 as positive controls for CTD-S2P binding, respectively. Proteins were detected by Western blotting against CBP. A representative experiment is shown.
Mud2 interacts with Prp19C and is needed for Prp19C and TREX occupancy
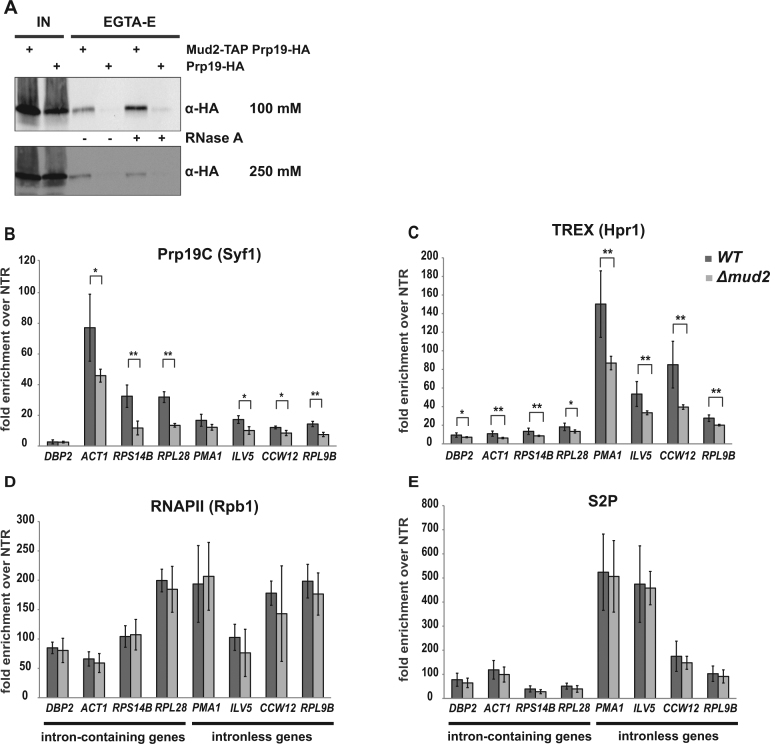
The decrease of both Mud2 and Prp19C occupancy in the S2A mutant suggests that Mud2 recruits Prp19C to the gene or vice versa. To assess whether Mud2 interacts physically with Prp19C in S. cerevisiae, we purified TAP-tagged Mud2 by tandem affinity purification (TAP). Mud2 was detected in Coomassie stainable amounts in the EGTA-eluate (Supplementary Figure S2A). Proteins co-purifying with Mud2 were identified by mass spectrometry (Supplementary Figure S2B). To determine whether Prp19C copurified with Mud2, we performed Western blots to detect endogenous HA-tagged Prp19 (Figure 4A). Indeed, Prp19 and thus most likely the whole Prp19 complex copurifies with Mud2 in vivo (Figure 4A, lane 3, upper panel), whereas it is absent from the negative control, TAP of a strain expressing HA-tagged Prp19 but non-tagged Mud2 (Figure 4A, lane 4, upper panel). This is consistent with previous findings that U2AF65, the potential human homolog of Mud2, interacts with PRP19C in human cells and that the Prp19C component Syf3 interacts with Mud2 in S. cerevisiae (41,66). The interaction between Mud2 and Prp19C is stable under higher salt concentrations, as Mud2 and Prp19 still copurify when the buffer contains 250 mM NaCl (Figure 4A, lane 3, negative control: lane 4, lower panel). Importantly, the Mud2-Prp19C interaction is independent of RNA as Prp19 copurifies with Mud2 when the extract is treated with RNaseA (Figure 4A, lane 5; negative control: lane 6). In contrast, RNaseA treatment abrogates or greatly decreases copurification of the nuclear mRNA-binding proteins Nab2 and Npl3 with Mud2 (Supplementary Figure S2A, lower panel). Thus, Mud2 and Prp19C interact in an RNA-independent manner in vivo.
Figure 4.
Mud2 interacts with Prp19C in vivo and is required for Prp19C and TREX occupancy. (A) Mud2-TAP was purified by tandem affinity purification (TAP). Copurification of the Prp19C subunit Prp19 with Mud2 in the EGTA eluate (EGTA-E) was assessed by Western blotting against the HA-tag on Prp19 (α-HA). TAPs were performed with standard (100 mM NaCl) and increased (250 mM NaCl) salt concentrations, and the RNA dependence of the interaction was tested by addition of RNase (+ RNaseA). A non-TAP-tagged PRP19-HA strain served as negative control. IN: input. (B) The occupancy of the Prp19C component Syf1 decreases in the Δmud2 mutant. Syf1 occupancy was analyzed at the 3′ ends of four exemplary intron-containing and four exemplary intronless genes by ChIP as in Figure 1B and C. (C–E) The occupancy of the TREX subunit Hpr1 decreases in the Δmud2 mutant, whereas the occupancy of the RNAPII subunit Rpb1 and the level of S2 phosphorylation are not affected. ChIP experiments as in (B).
To test whether Mud2 is needed for recruitment of Prp19C to the gene, we performed ChIP experiments with the Prp19C subunit Syf1 in cells deleted for MUD2 (Δmud2). Syf1 occupancy decreases at four exemplary intron-containing and four exemplary intronless genes in Δmud2 cells compared to wild-type cells (Figure 4B). Previously, we showed that Prp19C is required for TREX complex occupancy (29). Thus, TREX occupancy should also decrease in Δmud2 cells. As expected, the occupancy of the TREX component Hpr1 decreases in Δmud2 cells (Figure 4C). The decrease in Prp19C and TREX occupancy in Δmud2 cells is not caused by a decrease in the amount RNAPII and thus the CTD at the gene, as the occupancy of the RNAPII subunit Rpb1 does not change in Δmud2 cells compared to wild-type cells (Figure 4D). The decreased Prp19C and TREX occupancy is also not caused by a decrease in S2 phosphorylation as the latter stays constant in Δmud2 cells (Figure 4E). In terms of total cellular protein levels, deletion of MUD2 causes a slight increase in Syf1 levels and a slight decrease is Rpb1 levels but does not change Hpr1 levels (Supplementary Figure S2C). As total protein levels are largely unaffected in Δmud2 cells this is not the cause for the observed decrease in TREX and Prp19C occupancy. Furthermore, deletion of Mud2 specifically affects the recruitment of Prp19C and TREX to transcribed genes as the occupancy of the transcription elongation factors Paf1 and Spt5 does not change in Δmud2 cells (Supplementary Figure S3A and B). In addition, Prp19C is not needed for Mud2 occupancy as the occupancy of Mud2 is not decreased in syf1-37 cells, in which the partial C-terminal deletion of the Prp19C component Syf1 causes a decreased occupancy of Prp19C (29) (Supplementary Figure S3C). Likewise, TREX is not needed for Mud2 occupancy as Mud2 occupancy remains unchanged when the TREX component HPR1 is deleted (Supplementary Figure S3D). Thus, Mud2 acts ‘upstream’ of Prp19C and TREX. Taken together, Mud2 and Prp19C interact in vivo, and Mud2 is required for full occupancy of the Prp19 and TREX complexes.
The Mud2-Prp19C interaction and Mud2 occupancy at intronless genes are conserved in D. melanogaster
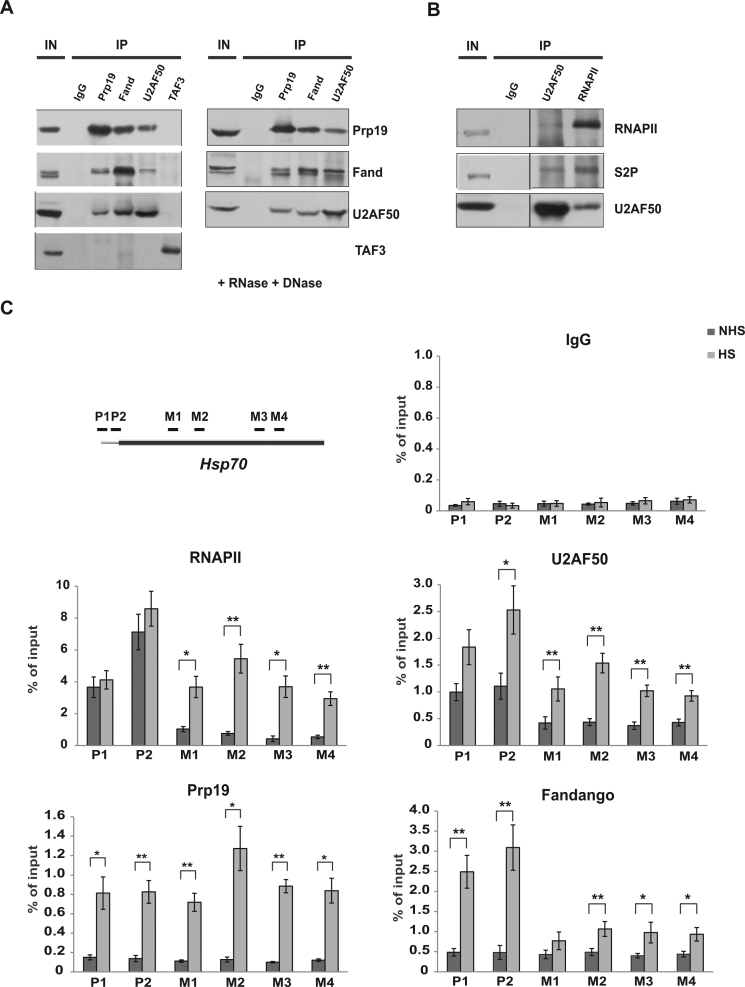
To assess whether the interaction of Mud2 with Prp19C is conserved in other organisms we chose Drosophila melanogaster as a model organism as it is one of the most elaborate model organisms among higher eukaryotes to study gene expression. To assess copurification of Mud2 and the Prp19 complex antibodies were raised against the Prp19C subunits Prp19 and Fandango (the D. melanogaster homolog of Syf1) and against U2AF50, the D. melanogaster homolog of Mud2 (Supplementary Figure S4A–C). Prp19 and Fandango coimmunoprecipitated each other from S2 cell extracts (Figure 5A, left panel) showing that also in D. melanogaster these two Prp19C subunits interact with each other. Importantly, Prp19 as well as Fandango also coimmunoprecipitated U2AF50 and, vice versa, U2AF50 coimmunoprecipitated Prp19 and Fandango (Figure 5A, left panel). In contrast, the unrelated protein TAF3, a subunit of the transcription initiation factor TFIID, does not interact with Prp19C components (Figure 5A, left panel). Furthermore, the interaction between Prp19C and Mud2 is independent of RNA and DNA since these proteins still coimmunoprecipitate when the extract is treated with RNase and DNase (Figure 5A, right panel, and Supplementary Figure S4D). Thus, Prp19C also interacts with Mud2 in Drosophila indicating that their function is also conserved.
Figure 5.
Mud2 interacts with Prp19C and is recruited to an intronless gene in D. melanogaster. (A) U2AF50, the Drosophila homolog of Mud2, interacts with the Prp19C complex components Prp19 and Fandango, the Drosophila homolog of Syf1. Prp19, Fandango or U2AF50 were immunoprecipitated with antibodies directed against the respective protein and (co-)purification in the immuno precipitate (IP) assessed by Western blotting (left panel). As negative control served TAF3, a subunit of the transcription initiation factor TFIID (left panel). The interactions between U2AF50, Prp19 and Fandango are independent of RNA and DNA as these proteins also copurify, when the extract is treated with RNase and DNase (right panel). (B) U2AF50 interacts with RNAPII in its S2 phosphorylated state in vivo. IP experiment and Western blots as in A using antibodies directed against RNAPII, the S2 phosphorylated CTD and U2AF50. (C) U2AF50, Prp19 and Fandango are recruited to the intronless Hsp70 gene in a transcription-dependent manner. ChIP experiments were performed with antibodies directed against U2AF50, Prp19, Fandango and RNAPII under non-induced (NHS: non-heat shock, dark-gray bars) and induced (HS: heat shock, light-gray bars) conditions. The position of the primer pairs used to determine the co-immunoprecipitated DNA at two positions in the promoter (P1, P2) and four positions in the ORF (M1–M4) of the Hsp70 gene are indicated in the top left panel.
In addition to the interaction between U2AF50 and Prp19C, we also assessed whether U2AF50 interacts with RNAPII in its S2 phosphorylated form in Drosophila. Indeed, U2AF50 copurifies RNAPII and RNAPII with an S2 phosphorylated CTD from Drosophila extracts (Figure 5B). Thus, the interaction between Mud2 and the S2 phosphorylated CTD of RNAPII is most likely conserved.
Next, we wanted to determine whether Prp19C and Mud2/U2AF50 are also recruited in a transcription-dependent manner to intronless genes in D. melanogaster. To do so, the occupancy of Prp19, Fandango, U2AF50 and RNAPII was assessed at the heat shock inducible Hsp70 gene. Under non-induced conditions, RNAPII is poised at the promoter, but absent from the gene body (Figure 5C, RNAPII, dark-gray bars). Prp19 and Fandango are absent from the promoter as well as the gene body, whereas U2AF50 is present at the promoter but absent from the gene body similar to RNAPII (Figure 5C, Prp19, Fandango, U2AF50, dark-gray bars). When expression of the Hsp70 gene is induced by heat shock, Hsp70 is transcribed as reflected by the increased occupancy of RNAPII at the promoter and throughout the ORF (Figure 5C, RNAPII, light-gray bars). In addition, the occupancies of Prp19, Fandango and U2AF50 increase significantly (Figure 5C, light-gray bars). Thus, the Prp19C and Mud2 are recruited to an intronless gene in a transcription-dependent manner in D. melanogaster. Taken together, the interaction of Prp19C and Mud2 and Mud2’s presence at intronless genes is conserved from yeast to Drosophila. Thus, the function of Mud2 in mRNA biogenesis is most likely conserved.
Mud2 functions in transcription
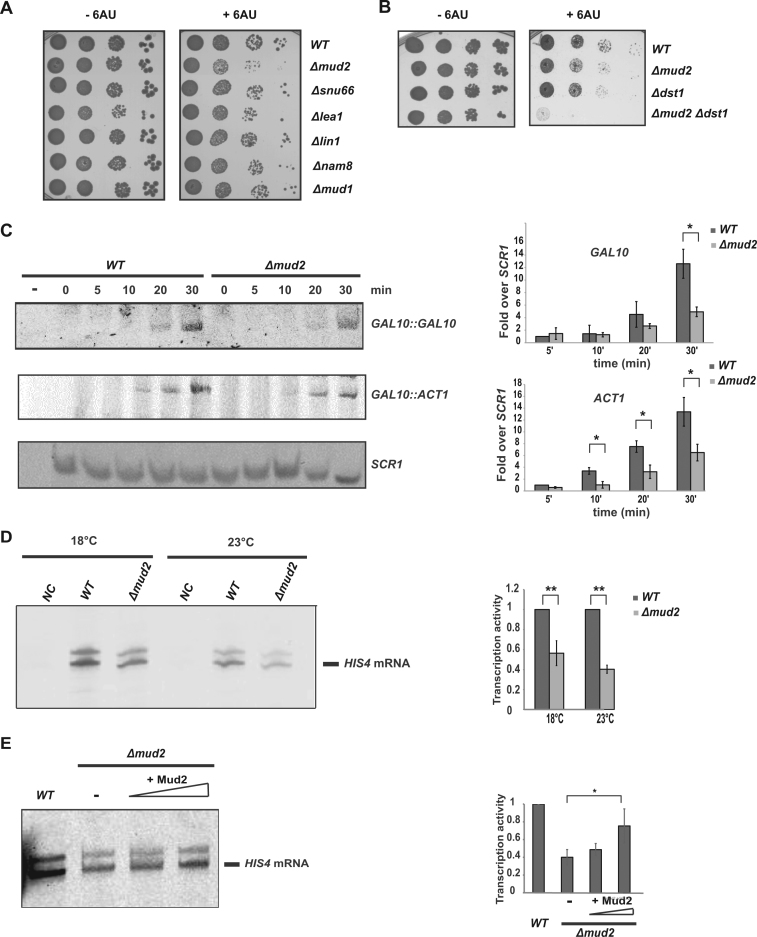
The interaction of Mud2 with Prp19C, which functions in transcription elongation, Mud2’s necessity for Prp19C occupancy and its presence at intronless genes indicate a function of Mud2 in transcription. To get a first indication for a function of Mud2 in transcription, we tested Δmud2 cells for sensitivity to 6-azauracil (6AU). 6AU decreases the intracellular pools of the nucleotides GTP and UTP and thus impairs transcription elongation. Cells deleted for MUD2 show sensitivity to 6AU (Figure 6A). This 6AU sensitivity of Δmud2 cells is most likely not a secondary effect of impaired splicing as cells deleted for the splicing factors Snu66 (component of the U4/U6.U5 snRNP), Lea1 (a component of U2 snRNP), Lin1 (a component of the U5 snRNP), Nam8 (a component of the U1 snRNP) or Mud1 (a component of the U1 snRNP) are not 6AU sensitive (Figure 6A). In contrast, the 6AU sensitivity of Δmud2 cells indicates that Mud2 is indeed needed for transcriptional activity possibly during elongation. The 6AU sensitivity caused by deletion of MUD2 is as strong as the one caused by deletion of DST1, the gene coding for the transcription elongation factor TFIIS (Figure 6B). Interestingly, deletion of both genes, MUD2 and DST1, causes a synthetically enhanced 6AU sensitivity (Figure 6B) providing genetic evidence that Mud2 functions in the same process as Dst1 but in a different manner. This is supported by the finding that deletion of MUD2 or deletion of DST1 cause a defect in co-transcriptional splicing, but at a different step of spliceosome assembly: by decreased recruitment of the U1 snRNP in Δmud2 cells and probably at a later stage of spliceosome assembly in Δdst1 cells (67). However, given the 6AU sensitivity of the Δmud2 strain this co-transcriptional splicing defect in both Δmud2 and Δdst1 cells could also reflect a defect in transcription elongation caused by deletion of MUD2.
Figure 6.
Mud2 functions in transcription. (A) Deletion of MUD2 causes 6AU sensitivity. Ten-fold serial dilutions of wild-type (WT, BY background), Δmud2, Δsnu66, Δlea1, Δlin1, Δnam8 and Δmud1 cells were spotted on SDC(-ura) plates containing solvent (–6AU) or 50 μg/ml 6AU (+6AU) and incubated for 2–3 days at 30°C. (B) Δmud2 is synthetically lethal with Δdst1 in the presence of 6AU. Ten-fold serial dilutions of wild-type (WT, BY background), Δmud2, Δdst1 and Δmud2 Δdst1 strains were spotted on SDC(-ura) plates containing solvent (–6AU) or 50 μg/ml 6AU (+6AU) and incubated for 2–3 days at 30°C. (C) Mud2 is required for efficient mRNA synthesis in vivo. Expression of the endogenous, intronless GAL10 gene and the plasmid-encoded, intron-containing ACT1 gene driven by the GAL10 promoter was induced for 5, 10, 20 and 30 min by the addition of galactose. Total RNA was extracted and the amount of GAL10 and ACT1 mRNA as well as the RNAPIII transcript SCR1, which served as loading control, determined by primer extension. One exemplary experiment is shown (left panel). The amount of GAL10 and ACT1 mRNA at the different time points of three independent experiments in wild-type (WT) and Δmud2 cells was quantified and normalized to the levels of the RNAPIII transcript SCR1. Values for wild-type cells 5 minutes after induction were set to 1 (right panel). (D) Deletion of MUD2 causes a transcription defect in vitro. The transcription activity of wild-type (WT) and Δmud2 cell extracts was assessed at 18 and 23°C by an in vitro transcription assay using a template containing a HIS4 promoter (left panel). The amount of synthesized RNA of three independent experiments is shown (right panel). (E) Add-back of recombinant Mud2 increases the transcription activity of a Δmud2 extract. In vitro transcription assay as in (D) with 50 and 300 ng of Mud2 added to the transcription reaction (left panel). The quantification of three independent experiments is shown (right panel).
Based on this 6AU sensitivity phenotype we could test, which domains of Mud2 are needed for its function (Supplementary Figure S5A). Thus, we constructed six partial deletion mutants of Mud2 (Supplementary Figure S5A). As only the deletion mutant that contains the two potential RRMs and the RRM of Mud2 complements the 6AU sensitivity of the Δmud2 deletion strain (Supplementary Figure S5B), only the less conserved N-terminal domain is dispensable for Mud2 function as assessed by this assay.
To obtain more direct evidence that Mud2 functions in transcription, we tested whether deletion of MUD2 affects transcription in vivo. To assess mRNA synthesis in vivo we used two reporter genes, both of which are driven by the galactose-inducible GAL10 promoter: the endogenous, intronless GAL10 gene and the intron-containing ACT1 gene. Δmud2 and wild-type cells were grown in raffinose-containing medium, and expression of both reporter genes was induced with galactose. RNA was isolated from both strains before induction and 5, 10, 20 and 30 min after induction, and the amount of synthesized GAL10 and ACT1 mRNA was quantified (Figure 6C, left panel). The levels of GAL10 and ACT1 mRNA are significantly decreased in the Δmud2 strain compared to the wild-type strain (Figure 6C, right panel). Thus, Mud2 is needed for the synthesis of normal levels of intron-containing and intronless mRNA in vivo.
To corroborate this in vivo finding and to determine whether the function of Mud2 in transcription is direct, we performed in vitro transcription experiments. To do this, we used nuclear extracts of wild-type and Δmud2 cells and a plasmid-based in vitro transcription assay with a HIS4 promoter (29). Δmud2 extracts had a significantly lower transcriptional activity than wild-type extracts at 18 and 23°C (Figure 6D). To assess whether the function of Mud2 in transcription suggested by this experiment is direct, we performed add-back experiments. For the add-back highly purified, recombinant Mud2 purified from E. coli or endogenous Mud2 purified from S. cerevisiae was used (Supplementary Figure S6A and B). Addition of increasing amounts of Mud2 to the in vitro transcription reaction consistently increased the transcriptional activity of the Δmud2 extract in a dose-dependent manner (recombinant Mud2: Figure 6E; Mud2 purified from S. cerevisiae: Supplementary Figure S6C). Importantly, RNAPII was not associated with Mud2 purified from S. cerevisiae under high salt conditions (Supplementary Figure S6D), and thus did not cause the stimulatory effect observed for addition of Mud2. The stimulatory effect of Mud2 was quite low indicating that the purified protein might be misfolded or that the extract of Δmud2 cells lacks another activity than the one of Mud2. Nevertheless, the above results show that Mud2 functions directly in transcription. Taken together, we show that Mud2 functions in S. cerevisiae to recruit the Prp19 and TREX complexes to the gene, which in turn is probably the underlying mechanism for Mud2’s function in transcription. Furthermore, this function of Mud2 is probably conserved in evolution.
DISCUSSION
The TREX complex plays crucial roles in transcription elongation, nuclear mRNP packaging and nuclear mRNA export. To fulfill these diverse functions and to couple transcription to nuclear mRNA export, TREX is already recruited to the transcribed gene in S. cerevisiae (13). Recruitment of TREX to the transcribed gene is mediated by its direct binding to the phosphorylated CTD (28). Second, TREX occupancy depends on the Prp19 splicing complex (29). The C-terminal domain of the Prp19C component Syf1 mediates the interaction between Prp19C and the transcription machinery (29). Here, we show that Mud2 is required for Prp19C and thus TREX occupancy in S. cerevisiae. Mud2 might recruit these two complexes to the mRNA based on the finding that Mud2 not only binds to intronic sequences of pre-mRNAs but also to intronless mRNAs in S. cerevisiae ((55) and Figure 1D–F). Interestingly, of all the splicing factors Msl5 has the highest ratio for binding to intronless transcripts versus intronic sequences of intron-containing transcripts (Figure 1F). Thus, Msl5 might also be involved in recruitment of Prp19C and TREX. In addition, Mud2 and Msl5 tend to cooccupy transcripts with Hpr1, Hrb1, Nab2 and Npl3 (55) further indicating that Mud2 and Msl5 might function in nuclear mRNP formation. Furthermore, U2AF50, the Drosophila homolog of the yeast Mud2, associates with transcripts derived from intronless genes and is necessary for the nuclear export of intronless mRNAs (68). In human, U2AF65, Prp19C and TREX bind to the so called CAR-E, an RNA consensus element that mediates the export of intronless mRNAs (69). Thus, Mud2 might not only recruit Prp19C and TREX to the transcribed gene but also to the mRNA.
Prp19C interacts with the transcription machinery via at least two interaction sites: Mud2 and the Prp19C subunit Syf1 (Figure 7). Mud2 interacts directly with the S2 phosphorylated CTD, whereas Prp19C interacts via the C-terminus of Syf1—directly or indirectly—with RNAPII (29). Consistent with this model, Prp19C occupancy decreases to 50% in Δmud2 cells as well as in syf1-37 cells, which lack the C-terminus of Syf1. Likewise, the occupancy of TREX most likely depends on at least two interaction sites: the CTD and Prp19C/Mud2 (Figure 7). Consistent with this model, there is also only a partial decrease in TREX occupancy in CTD mutants as well as in MUD2 or SYF1 mutants. Furthermore, as expected when Mud2 and Syf1 provide two independent anchor sites for TREX to the transcription machinery, combining the deletion of MUD2 with the syf1-37 mutation, which leads to reduced association of Prp19C with the transcription machinery, causes a synthetic lethal phenotype (Supplementary Figure S7). Interestingly, with the exception of an interaction of Mud2 with the cap-binding complex (70) all of these interactions ultimately require the CTD and are regulated by its phosphorylation, again pointing to the pivotal role of this coordinator in the transcription cycle (see introduction).
Figure 7.
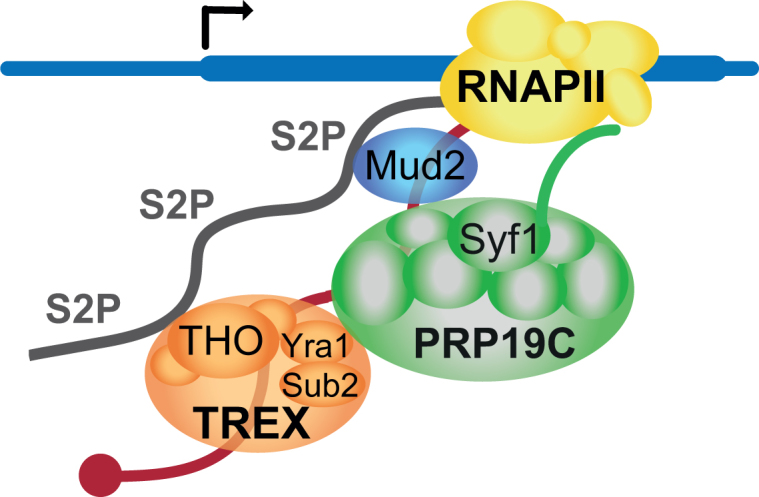
Model of Mud2’s functions in transcription and nuclear mRNP packaging. Mud2 is recruited to the transcription machinery by the S2 phosphorylated CTD. Mud2 also interacts with Prp19C and thus recruits this complex to the transcribed gene. Prp19C in turn is needed to maintain the occupancy of TREX, which also interacts directly with the S2 phosphorylated CTD. Mud2 is probably necessary for nuclear mRNP packaging as it recruits Prp19C and TREX to the mRNA. This in turn most likely causes the positive effect of Mud2 on transcription as the nascent mRNA is adequately bound by RNA-binding protein and thus does not hamper efficient transcription.
In contrast to U2AF50 in D. melanogaster, which is essential and necessary for the nuclear export of intronless mRNAs (68,71), Δmud2 cells do not show an mRNA export defect in S. cerevisiae (data not shown and (72)). This might be the reason why Mud2 is not essential in yeast. The absence of an obvious mRNA export defect in Δmud2 cells is consistent with the ∼50% occupancy of Prp19C and TREX at transcribed genes in Δmud2 cells compared to wild-type cells. Likewise, syf1-37 cells, which show a 50% occupancy of Prp19C and TREX, also do not exhibit an mRNA export defect (29). Thus, a lower amount of Prp19C and TREX at the transcribed gene is sufficient for nuclear mRNA export. This might at least partially be due to the fact that the rate of mRNA synthesis is also reduced in these mutant cells (also see below).
Both TREX and Prp19C are required for efficient transcription. Thus, consistent with a function in Prp19C and TREX occupancy, Mud2 is also required for full transcriptional activity as indicated by its sensitivity to 6AU and the decrease in transcriptional activity observed for Δmud2 cells in vitro and in vivo. Since the transcription defect of Δmud2 cells can be partially rescued by add-back of purified Mud2 the function of Mud2 in transcription is probably direct. Thus, we show for the first time that Mud2 is involved in transcription. During transcription, Mud2 most likely functions during transcription elongation as indicated by its 6AU sensitivity and its synthetically lethal phenotype when combined with deletion of DST1 in the presence of 6AU as well as its requirement for Prp19C and TREX occupancy since both complexes function in transcription elongation. As the complementation of the 6AU sensitive phenotype of Δmud2 cells requires the RRM and the two potential RRMs of Mud2 (Supplementary Figure S5), these three RRMs are probably needed for Mud2’s function in transcription. Due to impaired transcription elongation, total RNAPII levels slightly decrease in Δmud2 cells (Supplementary Figure S2C). This might be due to degradation of Rpb1 as shown for other mutants that impair transcription elongation (53). This decrease was neither observed for RNAPII occupancy nor S2 phosphorylation (Figure 2C and D), probably due to different sensitivities of the two methods or that the chromatin-bound fraction is not affected by the slight decrease in total RNAPII levels. Taken together, Mud2 most likely functions in transcription elongation by ensuring full Prp19C and TREX occupancy at the gene.
The function of Mud2 is probably largely conserved throughout evolution. In humans, U2AF65 interacts with the phosphorylated CTD and recruits Prp19C to the mRNA in in vitro splicing reactions, in which the CTD is tethered to the splicing substrate by an SRSF1-CTD fusion protein (41). Here, we show that in Drosophila U2AF50 interacts with Prp19C in vivo (Figure 5A). Furthermore, U2AF50 is recruited to the intronless Hsp70 gene in a transcription-dependent manner. Interestingly, U2AF50 stalls at the promoter similar to poised RNAPII under non-induced conditions suggesting that it interacts with poised RNAPII. Under induced conditions the profiles of Fandango and U2AF50 resemble the profile of RNAPII suggesting that Fandango ‘joins’ RNAPII at the promoter. In contrast, Prp19 shows a different profile (Figure 5C). Thus, similar to humans, which probably have three different Prp19C complexes, there might be different Prp19 complexes in Drosophila. It remains to be shown whether U2AF65/U2AF50—in humans, Drosophila and other higher eukaryotes—also functions in Prp19C as well as TREX recruitment and in transcription.
The different steps of gene expression are coupled. The bona fide function of Mud2 is in splicing (42–44). Here, we show that Mud2 is also necessary for Prp19C and TREX occupancy as well as efficient transcription. Since Mud2 is necessary for Prp19C and TREX occupancy, it likely also functions in nuclear mRNP packaging. Although, it needs to be determined whether Mud2 is involved in the coupling or coregulation of all or some of the processes it is involved in, Mud2 might coordinate transcription, splicing and nuclear mRNA export by recruitment of the two important protein complex Prp19C and TREX to the transcribed gene and the synthesized mRNA. Interestingly, deletion of MUD2 rescues the lethal phenotype of Δsub2 cells suggesting together with other findings that Sub2 functions to remove Mud2 during splicing (58). However, this genetic interaction might also reflect the functions of Mud2 and Sub2 in transcription and nuclear mRNP biogenesis. Furthermore, Mud2 interacts genetically and biochemically with Nab2, a nuclear mRNA binding protein involved in splicing, nuclear mRNP packaging and poly(A)tail length control (72,73). Furthermore, the vertebrate homolog of Nab2, ZC3H14, physically interacts with U2AF65 (72). The interaction between Nab2 and spliceosome components probably serves as a control mechanism to ensure that only properly processed mRNAs are exported out of the nucleus (72). As U2AF65 binds to the phosphorylated CTD and recruits Prp19C to the pre-mRNA, it was proposed to bridge transcription with spliceosome assembly (41). These findings further indicate that Mud2 might play a pivotal role not only in different nuclear steps of gene expression but also in the coordination of these steps.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Sittinan Chanarat (Mahidol University, Thailand) and Max Reuter (Imperial College London, UK) for critical reading of the manuscript, Susanne Röther, Silvia Hiechinger, Christoph Wierschem and Philipp Keil for generation of the RPB1 shuffle, SPT5-TAP, PAF1-TAP and MUD2-FTpA strains, respectively, Dirk Eick for antibody 3E10, Patrick Cramer (MPI, Göttingen) for plasmid LL279 and recombinant Gcn4. We are grateful to Günter Lochnit (Protein Analytics, Biochemical Institute, Justus Liebig University Giessen) for identification of copurifying proteins by mass spectrometry.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Research Foundation [STR697/2-1 and STR697/3-1 to K.S., GRK1591 to S.H.]; RFBR [16-34-01317 mol_a to V.P.]; Molecular and Cellular Biology program of the Russian Academy of Science [to D.K.]; program of fundamental research for state academies for 2013–2020 [01201363822 to S.G.]. Funding for open access charge: STR697/3-1 [to K.S.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Meinel D.M., Strasser K.. Co-transcriptional mRNP formation is coordinated within a molecular mRNP packaging station in S. cerevisiae. Bioessays. 2015; 37:666–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Singh G., Pratt G., Yeo G.W., Moore M.J.. The clothes make the mRNA: past and present trends in mRNP fashion. Annu. Rev. Biochem. 2015; 84:325–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Muller-McNicoll M., Neugebauer K.M.. How cells get the message: dynamic assembly and function of mRNA-protein complexes. Nat. Rev. Genet. 2013; 14:275–287. [DOI] [PubMed] [Google Scholar]
- 4. Katahira J. Nuclear export of messenger RNA. Genes (Basel). 2015; 6:163–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bjork P., Wieslander L.. Integration of mRNP formation and export. Cell. Mol. Life Sci.: CMLS. 2017; 74:2875–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mitchell S.F., Parker R.. Principles and properties of eukaryotic mRNPs. Mol. Cell. 2014; 54:547–558. [DOI] [PubMed] [Google Scholar]
- 7. Kurshakova M.M., Georgieva S.G., Kopytova D.V.. [Protein complexes coordinating mRNA export from the nucleus into the cytoplasm]. Mol. Biol. (Mosk). 2016; 50:723–729. [DOI] [PubMed] [Google Scholar]
- 8. Zhang D.W., Rodriguez-Molina J.B., Tietjen J.R., Nemec C.M., Ansari A.Z.. Emerging views on the CTD code. Genet. Res. Int. 2012; 2012:347214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Harlen K.M., Churchman L.S.. The code and beyond: transcription regulation by the RNA polymerase II carboxy-terminal domain. Nat. Rev. Mol. Cell Biol. 2017; 18:263–273. [DOI] [PubMed] [Google Scholar]
- 10. Tietjen J.R., Zhang D.W., Rodriguez-Molina J.B., White B.E., Akhtar M.S., Heidemann M., Li X., Chapman R.D., Shokat K., Keles S. et al. . Chemical-genomic dissection of the CTD code. Nat. Struct. Mol. Biol. 2010; 17:1154–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Suh H., Ficarro S.B., Kang U.B., Chun Y., Marto J.A., Buratowski S.. Direct analysis of phosphorylation sites on the Rpb1 C-Terminal domain of RNA polymerase II. Mol. Cell. 2016; 61:297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schuller R., Forne I., Straub T., Schreieck A., Texier Y., Shah N., Decker T.M., Cramer P., Imhof A., Eick D.. Heptad-specific phosphorylation of RNA polymerase II CTD. Mol. Cell. 2016; 61:305–314. [DOI] [PubMed] [Google Scholar]
- 13. Strasser K., Masuda S., Mason P., Pfannstiel J., Oppizzi M., Rodriguez-Navarro S., Rondon A.G., Aguilera A., Struhl K., Reed R. et al. . TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002; 417:304–308. [DOI] [PubMed] [Google Scholar]
- 14. Hurt E., Luo M.J., Rother S., Reed R., Strasser K.. Cotranscriptional recruitment of the serine-arginine-rich (SR)-like proteins Gbp2 and Hrb1 to nascent mRNA via the TREX complex. PNAS. 2004; 101:1858–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Heath C.G., Viphakone N., Wilson S.A.. The role of TREX in gene expression and disease. Biochem. J. 2016; 473:2911–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Luna R., Rondon A.G., Aguilera A.. New clues to understand the role of THO and other functionally related factors in mRNP biogenesis. Biochim. Biophys. Acta. 2012; 1819:514–520. [DOI] [PubMed] [Google Scholar]
- 17. Rondon A.G., Jimeno S., Garcia-Rubio M., Aguilera A.. Molecular evidence that the eukaryotic THO/TREX complex is required for efficient transcription elongation. J. Biol. Chem. 2003; 278:39037–39043. [DOI] [PubMed] [Google Scholar]
- 18. Strasser K., Hurt E.. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 2000; 19:410–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Strasser K., Hurt E.. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature. 2001; 413:648–652. [DOI] [PubMed] [Google Scholar]
- 20. Gwizdek C., Iglesias N., Rodriguez M.S., Ossareh-Nazari B., Hobeika M., Divita G., Stutz F., Dargemont C.. Ubiquitin-associated domain of Mex67 synchronizes recruitment of the mRNA export machinery with transcription. PNAS. 2006; 103:16376–16381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zenklusen D., Vinciguerra P., Wyss J.C., Stutz F.. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell. Biol. 2002; 22:8241–8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stewart M. Nuclear export of mRNA. Trends Biochem. Sci. 2010; 35:609–617. [DOI] [PubMed] [Google Scholar]
- 23. Jimeno S., Rondon A.G., Luna R., Aguilera A.. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J. 2002; 21:3526–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gaillard H., Wellinger R.E., Aguilera A.. A new connection of mRNP biogenesis and export with transcription-coupled repair. Nucleic Acids Res. 2007; 35:3893–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rougemaille M., Dieppois G., Kisseleva-Romanova E., Gudipati R.K., Lemoine S., Blugeon C., Boulay J., Jensen T.H., Stutz F., Devaux F. et al. . THO/Sub2p functions to coordinate 3′-end processing with gene-nuclear pore association. Cell. 2008; 135:308–321. [DOI] [PubMed] [Google Scholar]
- 26. Johnson S.A., Cubberley G., Bentley D.L.. Cotranscriptional recruitment of the mRNA export factor Yra1 by direct interaction with the 3′ end processing factor Pcf11. Mol. Cell. 2009; 33:215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. MacKellar A.L., Greenleaf A.L.. Cotranscriptional association of mRNA export factor Yra1 with C-terminal domain of RNA polymerase II. J. Biol. Chem. 2011; 286:36385–36395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meinel D.M., Burkert-Kautzsch C., Kieser A., O’Duibhir E., Siebert M., Mayer A., Cramer P., Soding J., Holstege F.C., Strasser K.. Recruitment of TREX to the transcription machinery by its direct binding to the phospho-CTD of RNA polymerase II. PLos Genet. 2013; 9:e1003914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chanarat S., Seizl M., Strasser K.. The Prp19 complex is a novel transcription elongation factor required for TREX occupancy at transcribed genes. Genes Dev. 2011; 25:1147–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chanarat S., Burkert-Kautzsch C., Meinel D.M., Strasser K.. Prp19C and TREX: interacting to promote transcription elongationand mRNA export. Transcription. 2012; 3:8–12. [DOI] [PubMed] [Google Scholar]
- 31. Chanarat S., Strasser K.. Splicing and beyond: the many faces of the Prp19 complex. Biochim. Biophys. Acta. 2013; 1833:2126–2134. [DOI] [PubMed] [Google Scholar]
- 32. Hogg R., McGrail J.C., O’Keefe R.T.. The function of the NineTeen Complex (NTC) in regulating spliceosome conformations and fidelity during pre-mRNA splicing. Biochem. Soc. Trans. 2010; 38:1110–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tran D.D., Saran S., Koch A., Tamura T.. mRNA export protein THOC5 as a tool for identification of target genes for cancer therapy. Cancer Lett. 2016; 373:222–226. [DOI] [PubMed] [Google Scholar]
- 34. Rehwinkel J., Herold A., Gari K., Kocher T., Rode M., Ciccarelli F.L., Wilm M., Izaurralde E.. Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat. Struct. Mol. Biol. 2004; 11:558–566. [DOI] [PubMed] [Google Scholar]
- 35. Herold N., Will C.L., Wolf E., Kastner B., Urlaub H., Luhrmann R.. Conservation of the protein composition and electron microscopy structure of Drosophila melanogaster and human spliceosomal complexes. Mol. Cell. Biol. 2009; 29:281–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Makarova O.V., Makarov E.M., Urlaub H., Will C.L., Gentzel M., Wilm M., Luhrmann R.. A subset of human 35S U5 proteins, including Prp19, function prior to catalytic step 1 of splicing. EMBO J. 2004; 23:2381–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kuraoka I., Ito S., Wada T., Hayashida M., Lee L., Saijo M., Nakatsu Y., Matsumoto M., Matsunaga T., Handa H. et al. . Isolation of XAB2 complex involved in pre-mRNA splicing, transcription, and transcription-coupled repair. J. Biol. Chem. 2008; 283:940–950. [DOI] [PubMed] [Google Scholar]
- 38. Guilgur L.G., Prudencio P., Sobral D., Liszekova D., Rosa A., Martinho R.G.. Requirement for highly efficient pre-mRNA splicing during Drosophila early embryonic development. eLife. 2014; 3:e02181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chan S.P., Kao D.I., Tsai W.Y., Cheng S.C.. The Prp19p-associated complex in spliceosome activation. Science. 2003; 302:279–282. [DOI] [PubMed] [Google Scholar]
- 40. Chung S., Zhou Z., Huddleston K.A., Harrison D.A., Reed R., Coleman T.A., Rymond B.C.. Crooked neck is a component of the human spliceosome and implicated in the splicing process. Biochim. Biophys. Acta. 2002; 1576:287–297. [DOI] [PubMed] [Google Scholar]
- 41. David C.J., Boyne A.R., Millhouse S.R., Manley J.L.. The RNA polymerase II C-terminal domain promotes splicing activation through recruitment of a U2AF65-Prp19 complex. Genes Dev. 2011; 25:972–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Abovich N., Rosbash M.. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997; 89:403–412. [DOI] [PubMed] [Google Scholar]
- 43. Wang Q., Zhang L., Lynn B., Rymond B.C.. A BBP-Mud2p heterodimer mediates branchpoint recognition and influences splicing substrate abundance in budding yeast. Nucleic Acids Res. 2008; 36:2787–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Berglund J.A., Chua K., Abovich N., Reed R., Rosbash M.. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell. 1997; 89:781–787. [DOI] [PubMed] [Google Scholar]
- 45. Rother S., Burkert C., Brunger K.M., Mayer A., Kieser A., Strasser K.. Nucleocytoplasmic shuttling of the La motif-containing protein Sro9 might link its nuclear and cytoplasmic functions. RNA. 2010; 16:1393–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chapman R.D., Heidemann M., Albert T.K., Mailhammer R., Flatley A., Meisterernst M., Kremmer E., Eick D.. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science. 2007; 318:1780–1782. [DOI] [PubMed] [Google Scholar]
- 47. Boehm A.K., Saunders A., Werner J., Lis J.T.. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol. Cell. Biol. 2003; 23:7628–7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Seraphin B.. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001; 24:218–229. [DOI] [PubMed] [Google Scholar]
- 49. Kornprobst M., Turk M., Kellner N., Cheng J., Flemming D., Kos-Braun I., Kos M., Thoms M., Berninghausen O., Beckmann R. et al. . Architecture of the 90S pre-ribosome: a structural view on the birth of the eukaryotic ribosome. Cell. 2016; 166:380–393. [DOI] [PubMed] [Google Scholar]
- 50. Kurshakova M.M., Krasnov A.N., Kopytova D.V., Shidlovskii Y.V., Nikolenko J.V., Nabirochkina E.N., Spehner D., Schultz P., Tora L., Georgieva S.G.. SAGA and a novel Drosophila export complex anchor efficient transcription and mRNA export to NPC. EMBO J. 2007; 26:4956–4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vorobyeva N.E., Soshnikova N.V., Nikolenko J.V., Kuzmina J.L., Nabirochkina E.N., Georgieva S.G., Shidlovskii Y.V.. Transcription coactivator SAYP combines chromatin remodeler Brahma and transcription initiation factor TFIID into a single supercomplex. PNAS. 2009; 106:11049–11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Georgieva S., Kirschner D.B., Jagla T., Nabirochkina E., Hanke S., Schenkel H., de Lorenzo C., Sinha P., Jagla K., Mechler B. et al. . Two novel Drosophila TAF(II)s have homology with human TAF(II)30 and are differentially regulated during development. Mol. Cell. Biol. 2000; 20:1639–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Karakasili E., Burkert-Kautzsch C., Kieser A., Strasser K.. Degradation of DNA damage-independently stalled RNA polymerase II is independent of the E3 ligase Elc1. Nucleic Acids Res. 2014; 42:10503–10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ranish J.A., Yudkovsky N., Hahn S.. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 1999; 13:49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Baejen C., Torkler P., Gressel S., Essig K., Soding J., Cramer P.. Transcriptome maps of mRNP biogenesis factors define pre-mRNA recognition. Mol. Cell. 2014; 55:745–757. [DOI] [PubMed] [Google Scholar]
- 56. Edgar R., Domrachev M., Lash A.E.. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002; 30:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cherry J.M., Hong E.L., Amundsen C., Balakrishnan R., Binkley G., Chan E.T., Christie K.R., Costanzo M.C., Dwight S.S., Engel S.R. et al. . Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 2012; 40:D700–D705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kistler A.L., Guthrie C.. Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for sub2, an essential spliceosomal ATPase. Genes Dev. 2001; 15:42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gornemann J., Kotovic K.M., Hujer K., Neugebauer K.M.. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol. Cell. 2005; 19:53–63. [DOI] [PubMed] [Google Scholar]
- 60. Moore M.J., Schwartzfarb E.M., Silver P.A., Yu M.C.. Differential recruitment of the splicing machinery during transcription predicts genome-wide patterns of mRNA splicing. Mol. Cell. 2006; 24:903–915. [DOI] [PubMed] [Google Scholar]
- 61. Zamore P.D., Green M.R.. Identification, purification, and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. PNAS. 1989; 86:9243–9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. West M.L., Corden J.L.. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics. 1995; 140:1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lunde B.M., Reichow S.L., Kim M., Suh H., Leeper T.C., Yang F., Mutschler H., Buratowski S., Meinhart A., Varani G.. Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain. Nat. Struct. Mol. Biol. 2010; 17:1195–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Licatalosi D.D., Geiger G., Minet M., Schroeder S., Cilli K., McNeil J.B., Bentley D.L.. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol. Cell. 2002; 9:1101–1111. [DOI] [PubMed] [Google Scholar]
- 65. Ujvari A., Luse D.S.. Newly Initiated RNA encounters a factor involved in splicing immediately upon emerging from within RNA polymerase II. J. Biol. Chem. 2004; 279:49773–49779. [DOI] [PubMed] [Google Scholar]
- 66. Chung S., McLean M.R., Rymond B.C.. Yeast ortholog of the Drosophila crooked neck protein promotes spliceosome assembly through stable U4/U6.U5 snRNP addition. RNA. 1999; 5:1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lacadie S.A., Tardiff D.F., Kadener S., Rosbash M.. In vivo commitment to yeast cotranscriptional splicing is sensitive to transcription elongation mutants. Genes Dev. 2006; 20:2055–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Blanchette M., Labourier E., Green R.E., Brenner S.E., Rio D.C.. Genome-wide analysis reveals an unexpected function for the Drosophila splicing factor U2AF50 in the nuclear export of intronless mRNAs. Mol. Cell. 2004; 14:775–786. [DOI] [PubMed] [Google Scholar]
- 69. Lei H., Zhai B., Yin S., Gygi S., Reed R.. Evidence that a consensus element found in naturally intronless mRNAs promotes mRNA export. Nucleic Acids Res. 2013; 41:2517–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fortes P., Kufel J., Fornerod M., Polycarpou-Schwarz M., Lafontaine D., Tollervey D., Mattaj I.W.. Genetic and physical interactions involving the yeast nuclear cap-binding complex. Mol. Cell. Biol. 1999; 19:6543–6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kanaar R., Roche S.E., Beall E.L., Green M.R., Rio D.C.. The conserved pre-mRNA splicing factor U2AF from Drosophila: requirement for viability. Science. 1993; 262:569–573. [DOI] [PubMed] [Google Scholar]
- 72. Soucek S., Zeng Y., Bellur D.L., Bergkessel M., Morris K.J., Deng Q., Duong D., Seyfried N.T., Guthrie C., Staley J.P. et al. . The Evolutionarily-conserved polyadenosine RNA binding protein, Nab2, cooperates with splicing machinery to regulate the fate of pre-mRNA. Mol. Cell. Biol. 2016; 36:2697–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Soucek S., Corbett A.H., Fasken M.B.. The long and the short of it: the role of the zinc finger polyadenosine RNA binding protein, Nab2, in control of poly(A) tail length. Biochim. Biophys. Acta. 2012; 1819:546–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.