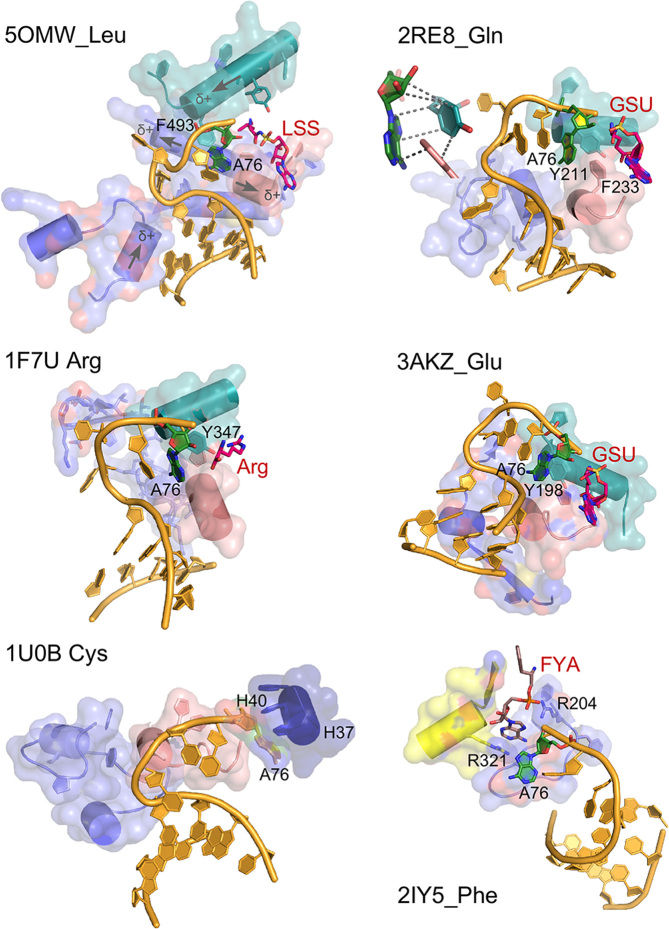
Figure 6.
AARS•tRNA complexes interacting from the minor groove side. PDB IDs and amino acids are as indicated. Active-site ligands are abbreviated as follows: LSS, 5′-O-(l-leucylsulfamoyl)adenosine; Arg, arginine; GSU, O5′-(l-glutamyl-sulfamoyl)-adenosine; FYA, adenosine-5′-[phenylalaninol-phosphate]. Class I secondary structures depicted by color include a homologous section of connecting peptide 1 (CP1), slate; the ‘specificity-determining helix’ teal; and the base of the helix from the second crossover of the Rossmann fold, containing the signature GxDQ, salmon), and in the case of PheRS the Motif-2 loop (slate) and Motif-3 (yellow). Electrostatic influences of the dipole moments of various helices are indicated on the helices of 5OMW_Leu. Side chains that interact with the tRNA 3′-terminal adenosine (A76) are indicated by number. Note that in the 1U0B (CysRS) complex the 3′-terminal adenosine occupies an unproductive position close to the HIGH catalytic signature, suggesting that in the absence of aminoacyl-5′-adenylate, the adenine ring finds a site similar to that normally occupied by the adenine ring of the adenylate, underscoring its potential binding affinity for the heterocycle. Inset in 2ER8_Gln shows the interaction between A76 and a conserved aromatic side chain at the N-terminus of the specificity-determining helix.