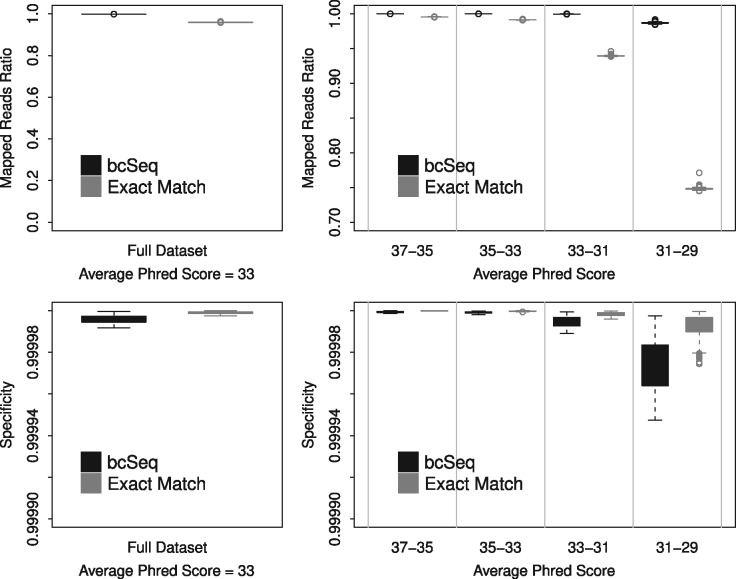
Fig. 1.
bcSeq maps more reads to barcodes for realistic data than perfect match without introducing significant mapping error. The upper left panel shows that bcSeq has a higher ratio of reads mapped to library barcodes (number of mapped reads/number of total reads) than perfect match when run on reads simulated using Phred scores sampled from real sequencing data. The upper right panel elucidates the role of sequencing error in bcSeq’s performance. Each read in the real sequencing data was binned based its average Phred score, and bcSeq and perfect match were run on reads simulated using Phred scores sampled from each of these bins. Perfect match’s mapped reads ratio degrades much more rapidly than bcSeq’s as average Phred score decreases. The lower panels compare the specificity (excluding unmapped reads) of bcSeq and perfect match. The lower left panel shows that bcSeq has a similar specificity to perfect match when run on reads simulated using Phred scores sampled from real sequencing data. The lower right panel elucidates the role of sequencing error in bcSeq’s specificity. bcSeq has a similar specificity to perfect match as the average Phred score decreases