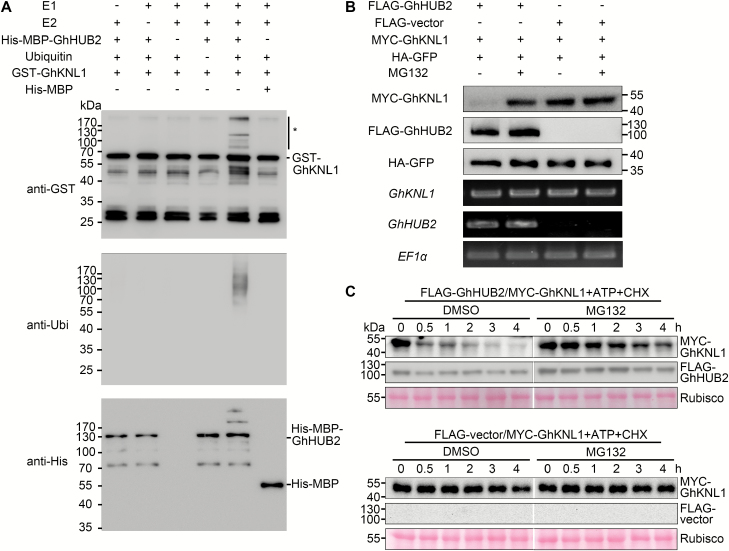
Fig. 6.
GhHUB2 ubiquitinates GhKNL1 and promotes the degradation of GhKNL1. (A) GhHUB2 ubiquitinates GhKNL1 in vitro. Ubiquitinated GST-GhKNL1 was detected by anti-GST and anti-Ubi antibodies. * indicates ubiquitinated GST-GhKNL1. (B) GhHUB2 promotes the degradation of GhKNL1 via the 26S proteasome in vivo. Immunoblotting analysis of protein extracts corresponding to agroinfiltrated Nicotiana benthamiana leaves with the indicated plasmids in the presence or absence of MG132. The abundance of MYC-GhKNL1 was detected using anti-MYC antibody and that of FLAG-GhHUB2 using anti-FLAG antibody. HA-GFP detection using anti-HA antibody served as a loading control. The mRNA expression levels of FLAG-GhHUB2 (GhHUB2) and MYC-GhKNL1 (GhKNL1) were analysed by RT-PCR; the mRNA expression level of EF1α was used as the internal control. (C) GhHUB2 promotes the degradation of GhKNL1 in a semi-in vivo protein degradation assay. FLAG-GhHUB2, MYC-GhKNL1, or FLAG vectors were individually expressed in N. benthamiana leaves for protein extraction. Extracts from MYC-GhKNL1 were mixed together with FLAG-GhHUB2 or FLAG vectors, after which the mixtures were treated with CHX and ATP in the presence of MG132 or DMSO for different times at 25 °C. At each time point, a portion of the mixture was removed and analysed by immunoblotting. The abundance of MYC-GhKNL1 was detected using anti-MYC antibody, and FLAG-GhHUB2 was detected using anti-FLAG antibody. Ponceau staining of Rubisco was used as a loading control.