FFig. 3.
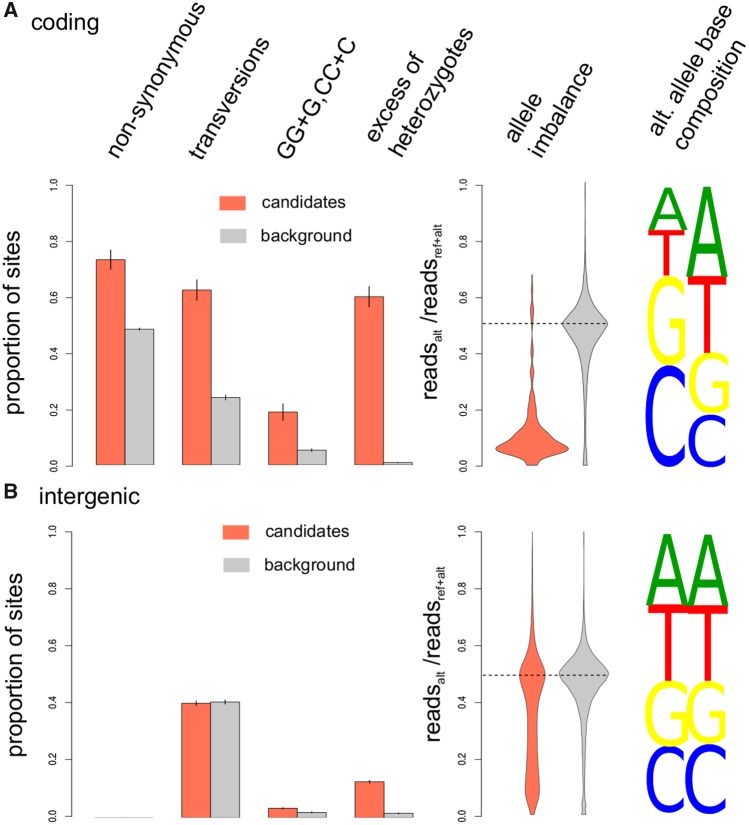
—Characteristics of error candidates in the 1000 Genomes data set detected in coding regions (a) or genome-wide based on the intergenic data set (b). For error candidates (red) and frequency-matched background variants (gray), the barplot shows the proportions of nonsynonymous versus synonymous variants, transversions versus transitions, alternative alleles introducing Gs or Cs after or before GG or CC dimers, and positions with significant excess of heterozygotes (P value < 0.05). The violin plot shows the proportion of sequences supporting the alternative alleles in individual with at least one sequence showing the alternative allele. On the right, the base composition of alternative alleles is shown for error candidates (red) and background variants (gray).