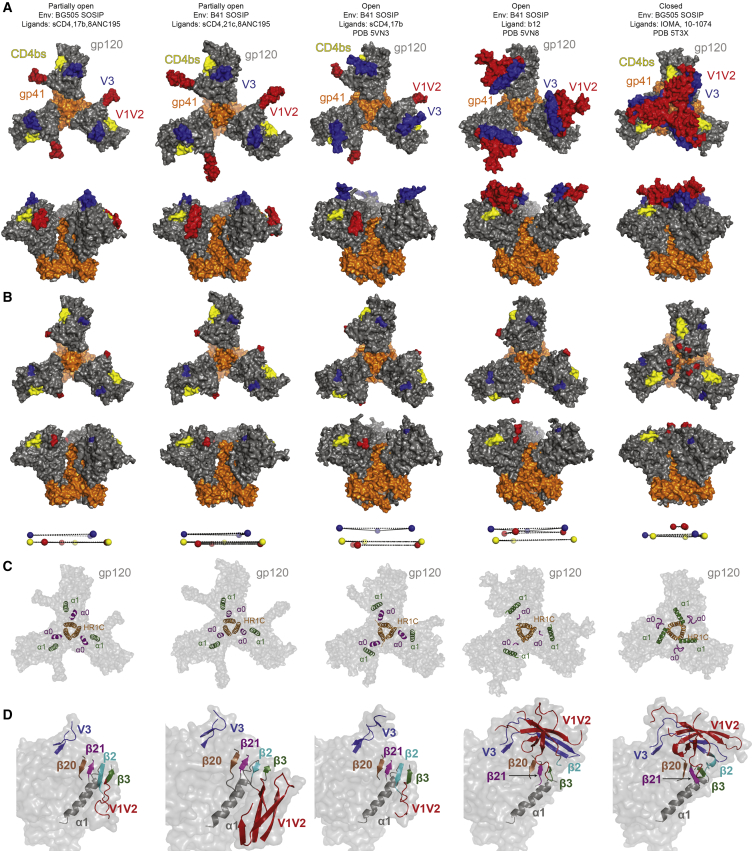
Figure 2.
Comparison of Env Conformations
(A) Top and side views of surface representations of Env trimers in partially open, sCD4-bound open, b12-bound open, and closed conformations. PDB: 5T3X is shown as a representative example of a closed Env structure; inter-protomer distances on closed Env structures are similar to each other despite strain-specific sequence differences and binding of different Fab ligands (Stadtmueller et al., 2018).
(B) Structures shown in (A) with coordinates for residues 126–198 in V1V2 and 296–330 in V3 removed to facilitate seeing positions of landmark residues: CD4bs residues 364–372 (yellow), residue 125 and 199 at the base of V1V2 (red), and residues 295 and 331 at the base of V3 (blue). Positions of CD4bs residue 368, V1V2 base residue 124, and V3 base residue 330 are shown as spheres below their respective Env structures.
(C) Conformational changes of α0. Structures in (A) and (B) are shown with coordinates for residues 64–73 in α0 and 98–117 in α1 in cartoon depiction overlaid on a transparent trimer surface.
(D) gp120 monomers from structures shown in (A) and (B). β2, β3, β20, β21, V1V2 residues 126–198, and V3 residues 296–330 are shown in cartoon representations overlaid on a transparent gp120 surface.