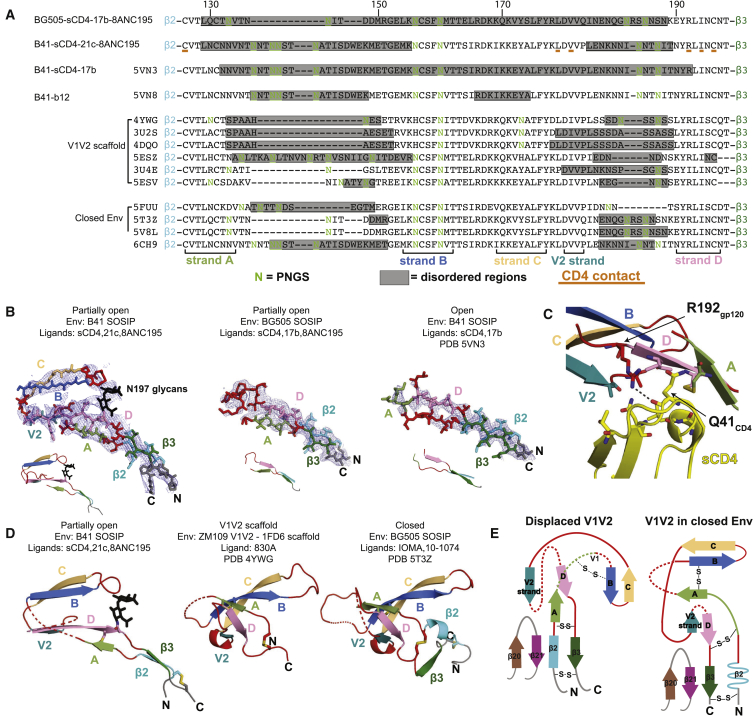
Figure 5.
V1V2 Conformations in Different Structures
(A) Sequence alignments of the V1V2 region from representative Env structures, including different conformations of Env trimers and monomeric V1V2 scaffolds. The A–D and V2 strands are indicated. Disordered regions in each structure are indicated by a gray background. Potential N-linked glycosylation sites (PNGSs) are highlighted in green and contacts with sCD4 (defined as residues in V1V2 with an atom within 5 Å of an sCD4 residue) in the B41-sCD4-21c-8ANC195 structure are underlined in red.
(B) Models of displaced V1V2 residues (stick representation) with accompanying density contoured at 8σ (0.047 e/Å3) for the indicated structures. Insets: cartoon representations of the V1V2 models.
(C) Close up of V1V2 interactions with sCD4 (yellow) in the B41-sCD4-21c-8ANC195 structure. A potential interaction with V1V2 residue Arg192gp120 and sCD4 is denoted as a black dashed line, and was identified based on distance.
(D) Cartoon representations of V1V2 regions in the indicated structures. Disordered regions are indicated by dashed lines.
(E) Topology diagrams of V1V2 structures shown in (D).