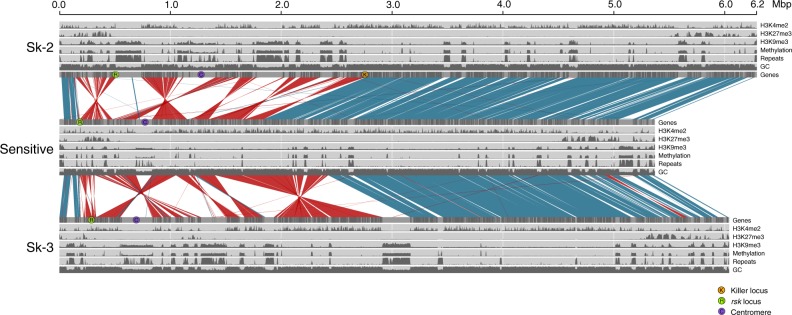
Fig. 1.
An alignment of chromosome 3 in Sk-2 and Sk-3 to sensitive Neurospora intermedia, which retains the ancestral gene order, shows a series of large inversions in the region between the killer and resistance loci. Blue color shows collinear regions and red inverted regions. All inversions are different between Sk-2 and Sk-3 and are interspersed with gene-poor and repeat-rich inserted regions. The location of the Sk-2 killer locus us marked with an orange circle and the rsk (resistance) locus and the centromeres are marked with green and purple circles, respectively. Tracks in gray show genes; GC content; repeat content; cytosine methylation; and H3K9me3, H3K27me3, and H3K4me2 histone methylation. H2K9me3 is a mark for constitutive heterochromatin, which in Neurospora is highly associated with repeats; H3K27me3 is a mark for facultative heterochromatin, which is primarily found in subtelomeric regions; and H3K4me2 is a mark for euchromatic regions. Cytosine methylation and the three different histone modifications were estimated using three biological replicates, and here the sum of the triplicates are presented