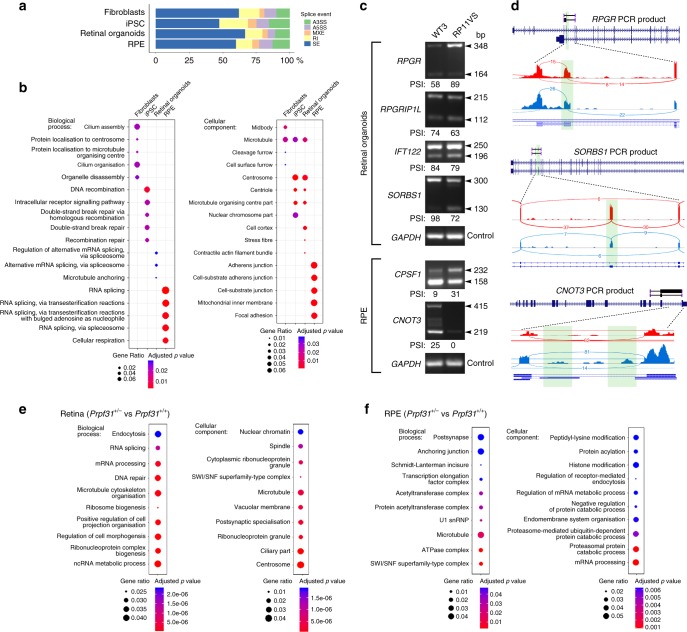
Fig. 4.
RNA-seq analysis of alternative splicing in fibroblasts, iPSC, RPE, retinal organoids and Prpf31+/− retina. a rMATS analysis showing that RP11 - RPE have the highest percentage of transcripts containing retained introns (RI) and alternative 3′ splice sites (A3SS); b Gene Ontology enrichment analysis showing biological and cellular processes affected by alternative splicing, respectively, in human cells; c Gel electrophoresis of RT-PCR for the indicated genes in RPE and retinal organoids derived from patient RP11VS and unaffected control WT3. Sizes (in bp) for major and minor isoforms (arrowheads), and percentage-spliced-in (PSI) values, are indicated; d Sashimi plots for the indicated genes for validation of alternative splicing events in RPE and retinal organoids derived from RP11 patients (blue) and unaffected controls (red). Data are representative of at least three independent experiments. Green highlights in Sashimi plots indicate alternative splicing events with the number of junction reads indicated for each event; e, f Gene Ontology enrichment analysis showing biological and cellular processes affected by alternative splicing, respectively, in mouse Prpf31+/− retinae and RPE. Data are representative of at least three independent experiments