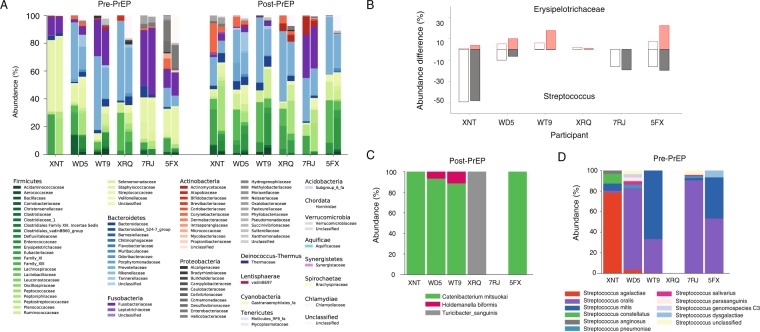
Figure 6.
Species identification using full-length 16S-23S sequencing. (A) Microbiome Family compositions measured by v4 region of 16s rRNA sequencing (left) and 16S-23S rRNA sequencing (right) for participants, XNT, WD5, WT9, XRQ, 7RJ, and 5FX at pre-PrEP and post-PrEP. (B) Abundance differences between pre-PrEP and post-PrEP specimens for Erysipelotrichaceae (red) and Streptococcus (grey) for study participants, XNT, WD5, WT9, XRQ, 7RJ, and 5FX. The abundance difference of Erysipelotrichaceae measured by 16S-23S sequencing (filled red boxes) was increased in four individuals, compared to the abundance difference obtained from v4-16s sequencing (empty red boxes). The abundance difference of Streptococcus measured by 16S-23S sequencing (filled grey boxes) was consistent with that measured by v4-16s sequencing (empty grey boxes). (C) Species composition of Erysipelotrichaceae family measured from participants XNT, WD5, WT9, XRQ, 7RJ, and 5FX after a minimum of 48 weeks PrEP. Within subjects XNT, WD5, WT9, and 5FX, Catenibacterium mitsuokai was the most dominant species in Erysipelotrichaceae family. Turicibacter sanguinis was detected in participant XRQ and Holdemanella biformis was detected in WD5 and WT9. (D) Species composition of Streptococcus genus measured from the above study participants before PrEP initiation. Streptococcus agalactiae (red) was most abundant in participant XNT, Streptococcus oralis (purple) was dominant in WD5, 7RJ and 5FX, and Streptococcus mitis (blue) was dominant in WT9.