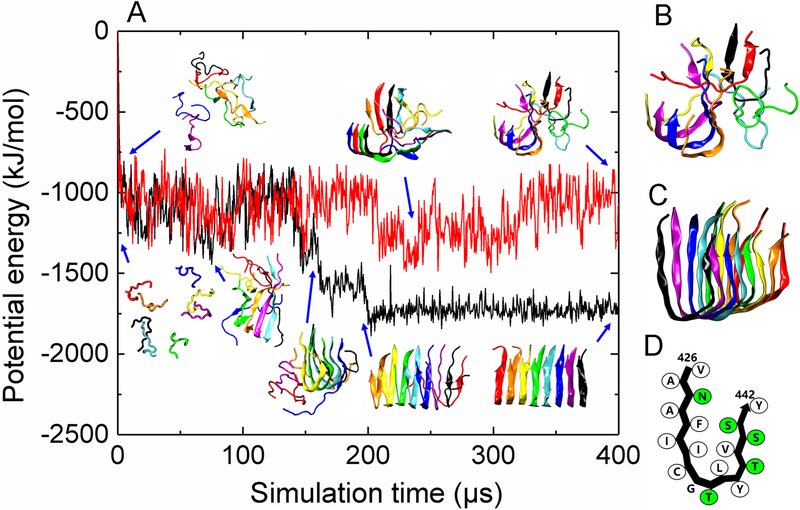
Figure 4.
DMD/PRIME20 simulations of P3. (A) A plot of potential energy of P3 peptide aggregation versus simulation time. Also shown are simulation snapshots of eight random coil P3 peptides aggregating to form a disordered oligomer (top panel corresponding to the red curve) and a U-shaped P3 protofilament (bottom panel corresponding to the black curve). Each of the eight peptides has a distinct color. (B) and (C) are final simulation snapshots of the disordered P3 oligomer and U-shaped P3 protofilament, respectively. (D) The schematic representation of peptide conformation in the U-shaped P3 protofilaments. Hydrophobic and polar residues are shown in white and green, respectively.