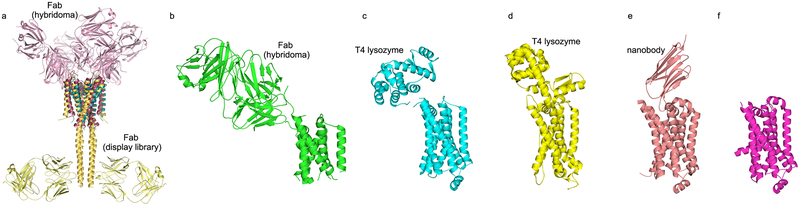
Figure 1. Comparison of selected membrane protein structures solved with and without chaperones.
(a) Superposition of KcsA structures. Green, structure solved without chaperones (PDB code 1BL8) [103]; pink, structure solved in high potassium salt, in complex with Fab obtained via hybridoma screening to recognize the functional KcsA tetramer (PDB code 1K4C)[68]; yellow, full-length KcsA structure solved in complex with Fab identified by molecular display technologies (PDB code 3EFF)[50]. (b) β2AR GPCR structure solved in complex with Fab fragment derived from hybridoma (PDB code 2R4R)[98]. (c-d) β2AR (PDB code 2RH1) [92] and A2AR (PDB code 3EML) [91] structures solved with covalent chaperone T4 lysozyme, respectively (e) β2AR structure solved in complex with nanobody (PDB code 3P0G) [104] (f) β1AR structure solved without chaperone assistance but including various modifications (PDB code 2VT4) [24]