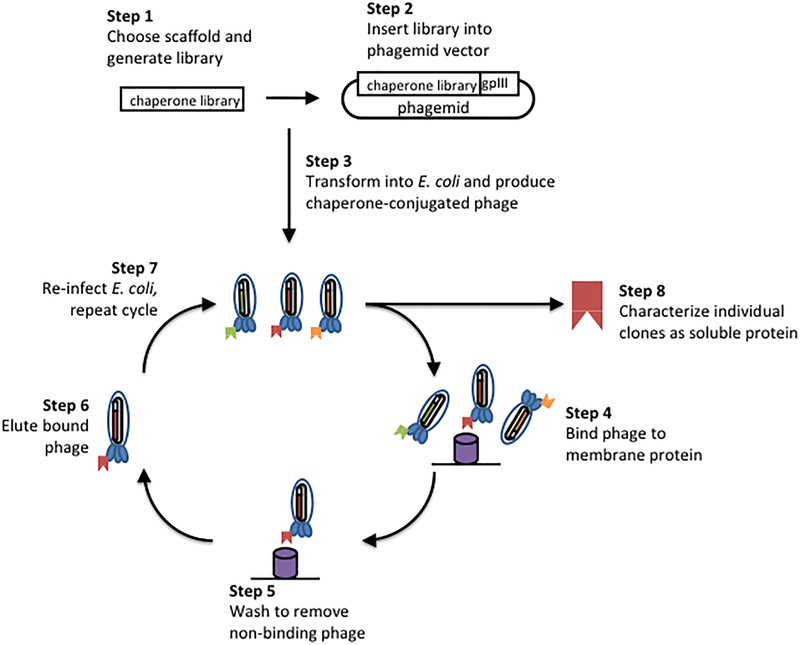
Figure 4. Phage display procedure for crystallization chaperone selection.
After selecting a chaperone and generating a library to engineer novel specificity, the chaperone library is inserted into a phagemid vector, which displays the chaperone as a fusion construct with a phage surface protein, such as the coat protein pIII. Phage expressed in E. coli contain the phagemid that encodes for the chaperone variant on the surface of the phage. The library of chaperone-conjugated phage is panned over the target membrane protein, and iterative rounds of binding, washing, elution and re-infection enrich the library for variants that bind the membrane protein. Hundreds of clones can be easily screened after multiple rounds of selection, and clones can be further optimized using additional rounds of phage display as desired.