Figure 3.
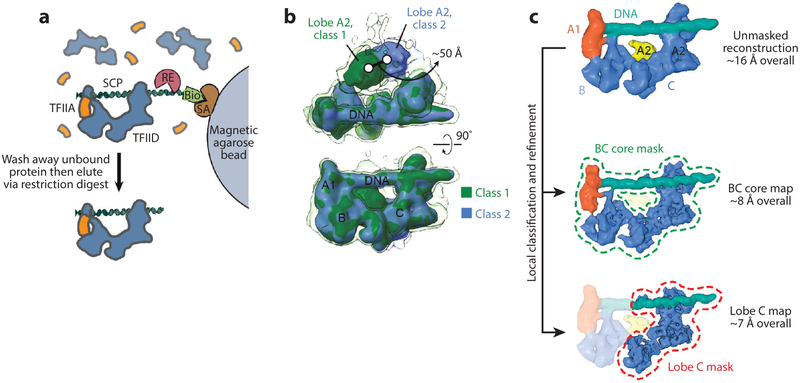
Cryo-electron microscopy (EM) analysis of promoter-bound TFIID. (a) Purification of promoter-bound TFIID-TFIIA complexes (52). Human TFIID complexes, previously affinity-purified from HeLa nuclear extract, are mixed with biotinylated (Bio) promoter DNA and recombinant human TFIIA (orange), and the DNA is then immobilized on magnetic streptavidin (SA) coated beads. Unbound DNA and proteins are washed away, and the purified DNA-bound complexes are then released by a restriction enzyme (RE) that cleaves the DNA at a restriction site placed between the promoter sequence and the biotinylated end. (b) 3 D classification of cryo-EM images of the TFIID-TFIIA-DNA complex reveals the flexibility of TFIID’s lobe A2 with respect to the more stable DNA-bound core composed of lobes A1, B, and C. The transparent isosurface is displayed at a lower threshold to enable visualization of weaker densities. (c) A strategy for local classification and refinement within masks around the DNA-bound core (dashed green line) or lobe C only (dashed red line) resulted in improved resolutions in these regions compared to the unmasked reconstruction (top) because of the exclusion of flexible parts of the complex. (Top, EMD 3304; middle, EMD 3305; bottom, EMD 3306.) Figure modified from References 52 and 60 with permission.