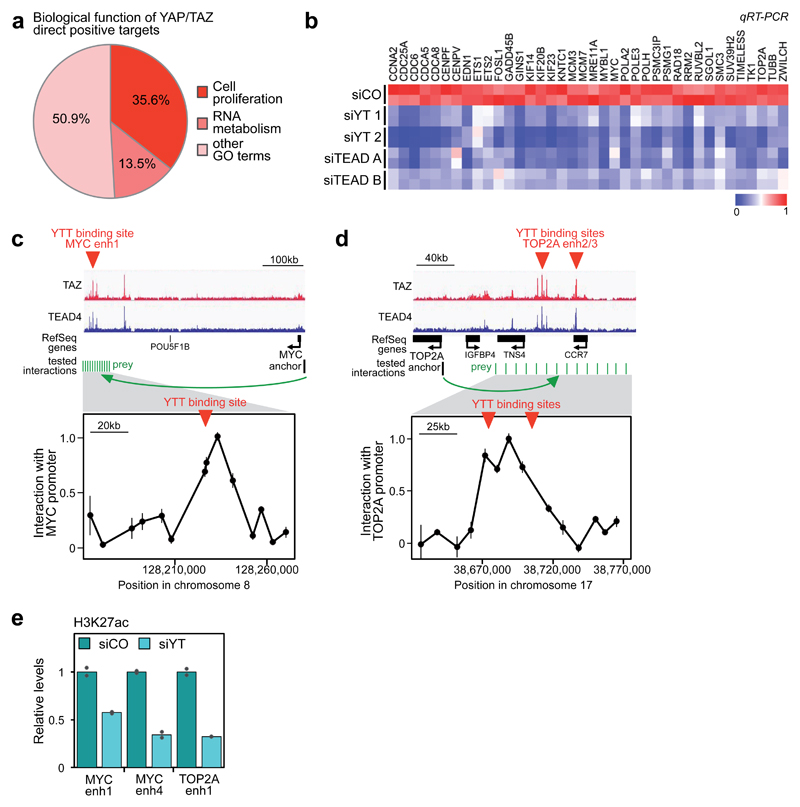
Figure 2. YAP/TAZ/TEAD transcriptional program.
(a) Biological functions associated to YAP/TAZ direct positive targets, identified by GO terms.
(b) YAP/TAZ or TEAD depletion impairs the expression of YAP/TAZ/TEAD direct target genes involved in cell proliferation, as evaluated by qRT-PCR (siCO=control siRNA, siYT=YAP/TAZ siRNA, siTEAD=TEAD siRNA). For a subset of genes, the downregulation of the corresponding proteins was also verified by Western blot (Supplementary Fig. 2b). See Supplementary Figure 2a for validation of TEAD siRNAs.
(c-d) Validation of the long-range interaction between YAP/TAZ-occupied enhancers and the promoters of MYC (c) and TOP2A (d) by DNA looping, using 3C assays in MDA-MB-231 cells. TAZ and TEAD4 ChIP-seq profiles show the position of YAP/TAZ/TEAD binding sites upstream of MYC or TOP2A genes (here named "MYC enhancer 1", “TOP2A enhancer2” “TOP2A enhancer3”), whereas no YAP/TAZ/TEAD binding sites were detected close to their TSS. The chart shows the frequency of interaction (measured as cross-linking frequency) between MYC or TOP2A promoter ("anchor") and the indicated sites surrounding YAP/TAZ/TEAD (YTT) peaks (green lines). Interaction frequency is higher close to YAP/TAZ peak. Data points are mean+SEM from n=3 biological replicates. See also Supplementary Figs. 2d-e for the additional interactions between MYC and TOP2A promoters and a different set of YAP/TAZ/TEAD-bound enhancers.
(e) ChIP-qPCR comparing the levels of H3K27ac (normalized to total H3 levels) in MDA-MB-231 cells transfected with control (siCO) or combined YAP/TAZ siRNAs (siYT1+2). Data from 2 biological replicates from one representative experiment are shown.
See Methods for reproducibility of experiments.