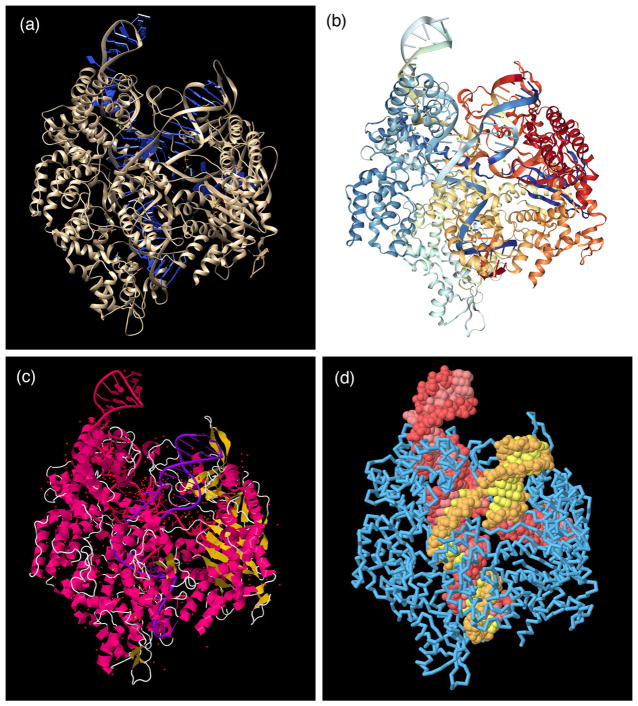
Figure 3.
The atomic structure of a Cas9 complex (PDB entry 4un3) displayed with several popular molecular graphics programs. (a) Chimera (hhtp://www.cgl.ucsf.edu/chimera) is designed for research and education, and by default presents an optimized view that highlights specific interactions between ligands (such as the small ion shown in green just right of center) and the macromolecules. (b) NGL (http://proteinformatics.charite.de/ngl) is the central molecular viewer at the RCSB PDB, by default providing a simplified introductory representation with rainbow-colored protein and nucleic acid chains and ball-and-stick for ligands and cofactors. (c) JSmol (http://jmol.sourceforge.net) is an easily scriptable program designed for embedding in websites. By default it provides a standard ribbon representation colored by the local secondary structure, and ball-and-stick for ligands, cofactors and all water molecules. (d) All of these programs provide many options for customizing the representation. For example, five minutes of scripting created this view in JSmol, highlighting the Cas9 protein (blue), the CRISPR RNA (red), and the targeted viral DNA (yellow). The protein is shown with a backbone trace to make the nucleic acid visible in the complex.