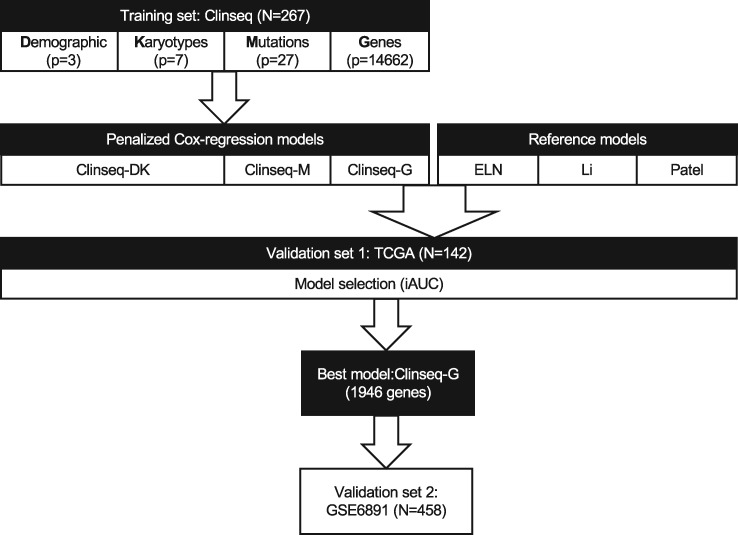
Figure 1.
Overall design of the study. The prediction model was developed in the training set, the Clinseq, with 267 acute myeloid leukemia (AML) patients. There are four types of predictors: D, the demographic parameters (age and sex); K, karyotypes (common cytogenetic aberrations in AML); M, mutations (27 somatic mutations); and G, genes (the normalized RNA-seq counts of 14 662 genes). Penalized (elastic-net) Cox regression models were fitted 1) Clinseq-DK, in the predictor sets D and K; Clinseq-M, in the predictor set M; Clinseq-G, in the predictor set G. Predictor sets D and K were combined to build one model. The prognostic scores of The Cancer Genome Atlas cohort were predicted by each model (Clinseq-DK, Clinseq-M, Clinseq-G). There are three models used as reference models: 1) the European LeukemiaNet risk classification system; 2) Li’s model based on a 24-gene expression signature; 3) Patel’s model based on an 18-gene panel of somatic mutations. The performance of the models was evaluated by the integrated area under the curve, integrated Brier score, concordance index, and hazard ratio of risk groups (dichotomized at the median prognostic score). The Clinseq-G model provided the best performance. In order to further investigate the generalizability, the Clinseq-G model was tested in the GSE6891 data set. ELN = European LeukemiaNet; iAUC = integrated area under the curve; TCGA = The Cancer Genome Atlas.