Abstract
Regulation of therapeutic transgene expression can increase the safety of gene therapy interventions, especially when targeting critical organs such as the brain. Although several gene expression systems have been described, none of the current systems has the required safety profile for clinical applications. Our group has previously adapted a system for novel gene regulation based on the destabilizing domain degron technology to successfully regulate glial cell-line derived neurotrophic factor in the brain (GDNF-F-DD). In the present study, we used GDNF-F-DD as a proof-of-principle molecule to fully characterize DD regulation in the brain. Our results indicate that DD could be regulated in a dose-dependent manner. In addition, GDNF-F-DD could also be induced in vivo repeatedly, without loss of activity or efficacy in vivo. Finally, DD regulation was able to be sustained for 24 weeks without loss of expression or any overt toxicity. The present study shows that DD has great potential to regulate gene expression in the brain.
Keywords: GDNF, destabilizing domains, in vivo, gene therapy, Parkinson’s disease
Graphical Abstract
Introduction
Gene therapy is experiencing a renaissance.1 Current viral vector technologies enable persistent expression of transgenes using in vivo and ex vivo gene delivery protocols. This has led to successful clinical trials that have paved the way for the market approval of gene therapy products in Europe to treat hereditary diseases.2 At a critical time when gene therapy transitions from the bench to the clinic, there are still key points to be addressed.
The first is efficacy, especially in disease-modifying approaches to treat complex diseases. For example, the neurturin (NTN) gene therapy clinical trials for Parkinson’s disease (PD) showed that the approach was safe both in phase 1 and phase 2 clinical trials, yet it failed to meet the primary endpoints at the phase 2 clinical trial.3, 4, 5 We now know that there was very limited distribution of NTN in the brain of patients. In addition, the patients had very advanced PD and may not have been responsive to a neurotrophic factor therapy that is aimed at rescuing the pathology at an earlier time point. With the emergence of better delivery methods6 and novel viral vectors with enhanced distribution,7 it will be possible to ensure that the target area is effectively transduced. Provided that a sufficient safety profile for gene therapy approaches can be established, it will soon be possible to design trials containing patient cohorts that are at an earlier disease stage and, therefore, more amenable to disease-modifying gene therapies.
Another hurdle to overcome is lack of regulation of transgene expression in transduced cells and tissues. Current viral vectors used in clinical trials are designed to have strong constitutive promoters resulting in long-term supraphysiological expression of transgenes. Once the viral vector is delivered, barring ablative surgery, it is not possible to adjust or stop transgene expression. This can be addressed by using an inducible system to regulate gene expression. However, none of the existing gene inducible systems has been approved for clinical applications.8
The destabilizing domain (DD) degron technology developed by the Wandless lab showed great promise in regulating gene expression in vivo.9, 10, 11 The DD gene-inducible system regulates gene expression at a post-translational level. The transgene of interest is expressed as a fusion protein together with a coding sequence for a DD. When the transgene-DD fusion is expressed, the DD moiety will target the full fusion protein to proteasomal degradation. In the DD variant we use, addition of trimethoprim (TMP) will block proteasomal recognition, thus allowing the protein of interest to be stabilized.
Our group has previously adapted the DD technology to regulate glial-cell-line-derived neurotrophic factor (GDNF).9, 10 The plethora of studies showing neuroprotective and neurorestorative effects of GDNF make this protein still one of the most promising therapeutic agents to treat PD.12, 13 However, constitutive high levels of GDNF expression in the brain can cause considerable side effects.14, 15 Thus, in order to avoid any side effects, GDNF expression needs to be localized and regulated, making it a very good candidate for preclinical testing of gene inducible systems.
We have previously shown that second-generation DD-regulated GDNF (GDNF-F-DD) was able to protect dopaminergic neurons when induced with TMP, and more importantly, no activation of GDNF signaling pathways was observed in the absence of TMP.10 Based on these promising results, we set out to perform preclinical validation of DD system regulation using GDNF-F-DD as a proof-of-principle molecule.
Results
TMP Regulates GDNF-F-DD in a Dose-Dependent Manner
One of the desired features of an inducible gene expression system, including post-translational systems such as DD, is the ability to respond in a dose-dependent manner to the controlling drug. To determine whether GDNF-F-DD stability could be regulated in a dose-dependent manner by TMP, lentiviral vectors (LVs) expressing GDNF or GDNF-F-DD were delivered bilaterally to the striatum of Sprague-Dawley (SD) female rats. One extra group of animals was kept as an untransduced control group. Three days after LV delivery, the GDNF-F-DD group was subdivided into four treatment groups receiving drinking water containing 0.05 mg/mL, 0.1 mg/mL, 0.2 mg/mL, or 0.5 mg/mL TMP continuously in the drinking water. One subgroup of GDNF-F-DD animals was given regular drinking water and was used as an uninduced GDNF-F-DD control (OFF) group. Three weeks after LV delivery, the striata of animals were dissected, and each hemisphere was counted as an independent data point.
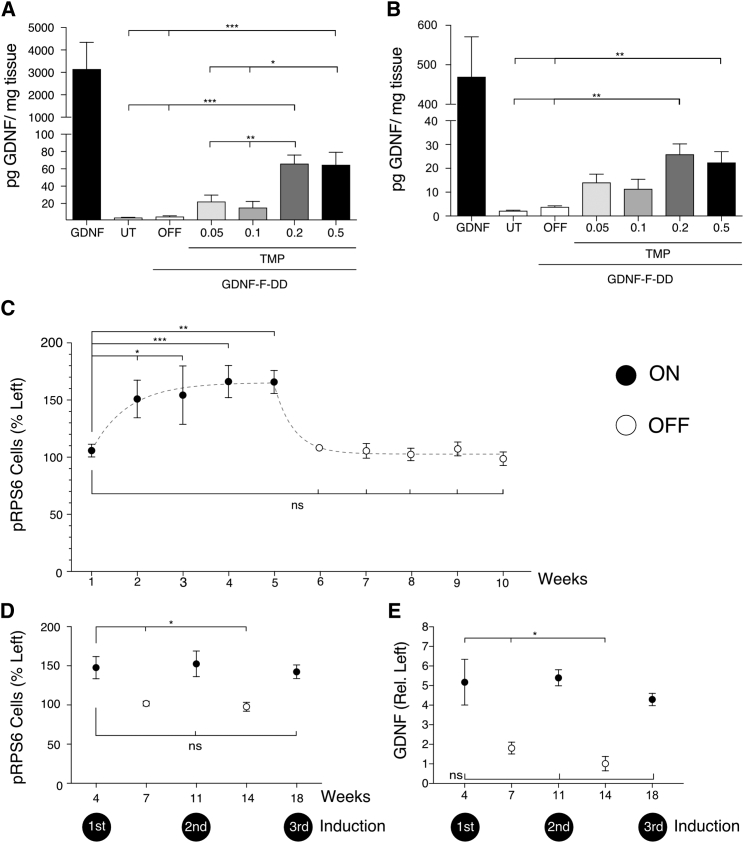
A GDNF ELISA of dissected striata (Figure 1A) indicated that wild-type GDNF used as a positive control was expressed at very high levels, reaching 3,153 ± 1,189 pg/mg. Untransduced animals had endogenous levels of GDNF averaging 3.7 ± 0.3 pg/mg, while the OFF animals had similar levels of GDNF, 5.0 ± 0.8 pg/mg. In the GDNF-F-DD animals receiving 0.05 mg/mL and 0.1 mg/mL TMP, the GDNF concentrations in the striatum increased to 22.4 ± 7.4 pg/mg and 15.45 ± 7.0 pg/mg, respectively. Induction of GDNF-F-DD reached a maximum concentration of 66.1 ± 9.8 pg/mg when the animals were given 0.2 mg/mL TMP. Giving animals 0.5 mg/mL TMP did not increase the GDNF concentration further, as in these animals, there was 64.8 ± 14.4 pg/mg of GDNF in the striatum. A one-way ANOVA of the untransduced and GDNF-F-DD groups indicated that there was a difference between groups, F = 12.04, p < 0.0001. Post hoc analysis with a Tukey multiple comparison test indicated that there was no significant GDNF increase in the groups that received 0.05 mg/mL TMP and 0.1 mg/mL TMP when compared with the untransduced and OFF groups. However, there was a significant increase of GDNF concentration in the groups that received 0.2 and 0.5 mg/mL TMP when compared with the untransduced and OFF groups, 0.05 mg/mL TMP and 0.1 mg/mL TMP, respectively.
Figure 1.
GDNF-F-DD Can Be Regulated in a Dose-Dependent Manner, Revert Rapidly to Basal Levels, and Be Induced Multiple Times In Vivo without Loss of Efficiency
LVs expressing GDNF-F-DD were delivered to the striatum of animals. One group of animals was given LV expressing GDNF, and another group of animals was untransduced (UT). After 2 days, the animals were given 0.05, 0.1, 0.2, or 0.5 mg/mL TMP continuously in the drinking water for 3 weeks. (A) GDNF ELISA was performed on striatum samples. (B) GDNF ELISA was performed in substantia nigra samples. A one-way ANOVA with Dunnett multiple comparison tests was performed (n = 4–6 per group). To determine the kinetics of the DD system in vivo, LVs expressing GDNF-F-DD were delivered to the right striatum of SD rats. The animals were given TMP (ON) for up to 5 weeks, and then the TMP was withdrawn and the animals given regular drinking water (OFF) for another 5 weeks. (C) Histological analysis and quantification of pRPS6 neurons were performed to determine how fast the DD system could be turned on and off (n = 3–5 per group). A similar experimental setup was used to determine whether the DD system could be induced multiple times. After LV GDNF-F-DD delivery, animals were divided into five groups. The first group was ON for 4 weeks (first induction). The second was ON for 4 weeks and OFF for 3 weeks. The next group of animals was induced 2 times, ON-OFF-ON (second induction). The following group was ON-OFF-ON-OFF. The last group was induced 3 times, ON-OFF-ON-OFF-ON (third induction). (D) Quantification of activated SNpc cells was performed in sections stained for pRPS6 (n = 4–5 per group). (E) Analysis of GDNF-F-DD expression was done using striatal sections stained for GDNF (n = 4–5 per group). The x axis indicates weeks after LV delivery (see also Figure S1). A one-way ANOVA with Dunnett multiple comparison tests was performed. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001. ns, not significant. Error bars represent mean ± SEM or mean + SEM.
Similarly, a GDNF ELISA of the substantia nigra (SN) (Figure 1B) indicated that the wild-type GDNF control group had very high levels of GDNF, 469.9 ± 101.1 pg/mg. The untransduced group had a basal level of GDNF of 2.2 ± 0.2 pg/mg, whereas the OFF group had 3.9 ± 0.4 pg/mg GDNF. There was, however, an increase in GDNF expression in the GDNF-F-DD animals that were given TMP. The subgroups that were given 0.05 mg/mL TMP and 0.1 mg/mL TMP had indistinguishable levels of 14.1 ± 3.5 mg/mL and 11.4 ± 4.0 mg/mL GDNF, respectively. Similar to the striatum, GDNF-F-DD, induction peaked in the subgroup that was given 0.2 mg/mL TMP, where it reached 26.0 ± 4.3 pg/mL GDNF. The subgroup that received 0.5 mg/mL TMP reached a GDNF concentration of 22.5 ± 4.5 mg/mL.
A one-way ANOVA comparing the untransduced and different GDNF-F-DD subgroups indicated a significant difference, F = 7.6, p < 0.0001. Post hoc analysis using Tukey multiple comparison test indicated that there was a significant increase of GDNF when the untransduced or OFF animals were compared with the animals given 0.2 and 0.5 mg/mL TMP.
GDNF-F-DD Reverts Rapidly to Basal Levels and Can Be Induced Multiple Times In Vivo
The next step was to determine the kinetics of GDNF-F-DD stabilization. LVs expressing GDNF-F-DD were delivered to the right striatum of SD female rats. After LV delivery, the animals were randomized into groups, and all were given 0.2 mg/mL TMP in the drinking water. To assess the kinetics of GDNF-F-DD activation, groups of animals were euthanized 1, 2, 3, 4, and 5 weeks after LV delivery. To assess the kinetics of GDNF-F-DD deactivation, after 5 weeks of activation, animals were switched to regular drinking water and euthanized 1, 2, 3, 4, and 5 weeks after TMP withdrawal.
Brains of animals were processed for histology and stained for phosphorylated ribosomal S6 protein (pRPS6), a known marker for GDNF activity in substantia nigra pars compacta (SNpc) neurons.10, 16, 17 Histological analysis of pRPS6-positive cells (Figures 1C, S1A, and S1B) in the injected side was quantified using the contralateral side as control. pRPS6 cell numbers indicated that 1 week after the start of TMP treatment, pRPS6 was still at basal levels, 105.8 ± 5.7%. GDNF-F-DD activation became noticeable at weeks 2 and 3 after activation of the system, with pRPS6 cell numbers reaching 151 ± 16.5% and 151.3 ± 25.6%, respectively. GDNF-F-DD activation reached its peak between weeks 4 (166.2 ± 14%) and 5 (165.8 ± 10.1%) after TMP treatment. However, once TMP was withdrawn, pRPS6 cell numbers decreased to basal levels quickly. One week after deactivation, the number of pRPS6 cells was already down to 97.2 ± 2.5%. From there on, pRPS6-positive cell numbers were 108.4 ± 4.1%, 101.0 ± 6.8%, 107.0 ± 7.3%, and 106.9 ± 7.9% for weeks 2, 3, 4, and 5 after deactivation, respectively.
A one-way ANOVA of pRPS6 cell number indicated statistically significant differences, F = 7.7, p < 0.0001; and a post hoc analysis using Dunnett multiple comparisons between week 1 of activation and the remaining groups was then performed. There was a statistically significant increase in the number of pRPS6 cells in the left hemisphere from weeks 2–5 after activation. Interestingly, there was no statistically significant difference between week 1 of activation and weeks 1–5 after TMP withdrawal. These data suggest that the DD system has very rapid deactivation kinetics.
After knowing the induction kinetics for GDNF-F-DD, an experiment was designed to ascertain whether GDNF-F-DD was able to withstand multiple induction cycles. LVs expressing GDNF-F-DD were delivered to the right striatum of female SD rats. After LV injection, the animals were randomly distributed into five groups and given 0.2 mg/mL TMP. One group had one round of TMP induction (ON) as the animals were given TMP for 4 weeks. The second group was ON for 4 weeks, TMP was subsequently withdrawn, and the animals were given regular drinking water for 3 weeks (ON-OFF). The third group had 2 rounds of induction and one round of TMP withdrawal (ON-OFF-ON). The fourth group had 2 rounds of induction and 2 rounds of TMP withdrawal (ON-OFF-ON-OFF). Lastly, the fifth group had three rounds of induction and 2 rounds of TMP withdrawal (ON-OFF-ON-OFF-ON). After the end of their final induction or withdrawal period, each group was euthanized, and the brains were processed for histological analysis.
Analysis of pRPS6-positive cells in SNpc neurons (Figures 1D and S1C) indicated that there was a robust induction of GDNF-F-DD activity in all three induction cycles. In the ON group, there were 147.7 ± 14% pRPS6-positive cells; in the ON-OFF-ON group, there were 152.6 ± 16.4% pRPS6-positive cells; and in the ON-OFF-ON-OFF-ON group, there were 142.4 ± 8.7% pRPS6-positive cells. On the other hand, once TMP was withdrawn, there were only baseline levels of GDNF activity, with 101.8 ± 3.0 and 97.7 ± 5.7% pRPS6-positive cells for the ON-OFF and ON-OFF-ON-OFF groups, respectively. A one-way ANOVA indicated significant differences between groups, F = 5.6, p < 0.005. Dunnett post hoc analysis using the ON group as control indicated that there was a significant decrease in pRPS6-positive cells when compared to the ON-OFF and ON-OFF-ON-OFF groups. Important, no statistically significant differences were seen when the first induction group was compared with the second and third inductions.
GDNF expression was also analyzed histologically (Figures 1E and S1D) and quantified by measuring GDNF densitometry as relative to the left intact side. The GDNF in the ON group was increased 5.2 ± 1.2-fold, whereas in the ON-OFF group, it was 1.8 ± 0.3-fold, compared to the contralateral intact side. This difference was maintained in the ON-OFF-ON and ON-OFF-ON-OFF groups, with 5.4 ± 0.4-fold and 1.0 ± 0.4-fold, respectively. Finally, the expression of GDNF was reverted back to 4.3 ± 0.3-fold in the ON-OFF-ON-OFF-ON group. A one-way ANOVA again indicated differences between groups, F = 6.0, p < 0.003. A Dunnett post hoc analysis using the ON group as control indicated that there was a significant decrease in GDNF-F-DD expression when compared to the ON-OFF and ON-OFF-ON-OFF groups. No statistically significant differences were seen between the ON group, ON-OFF-ON group, and ON-OFF-ON-OFF-ON group.
This set of experiments showed that it took 3 to 4 weeks for GDNF-F-DD to reach maximum activity and only 1 week of TMP withdrawal for the activity of GDNF-F-DD to be completely turned off. The characterization of GDNF-F-DD induction kinetics showed that TMP was able to regulate GDNF-F-DD, maintaining its expression and activity throughout at least 3 full induction cycles.
Assaying GDNF-F-DD Immune Response and Neurotoxicity In Vivo
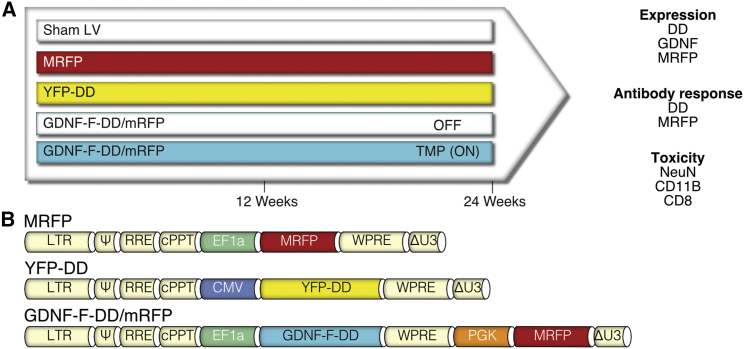
The last aspect to consider in an inducible system is the biosafety profile for both the activating molecule and the system itself. While TMP is a very well-characterized and safe drug,18 the biosafety of the DD system has never been evaluated in vivo. Studies using tetracycline inducible systems have shown that potential toxicity arises from an immune response against exogenous proteins from bacterial origin.19, 20 As the DD system also requires continuous translation of an exogenous bacterial protein, we designed an experiment to comprehensively evaluate long-term expression, immune responses, and potential neurotoxicity of GDNF-F-DD (Figure 2A).
Figure 2.
Experimental Design and LV Used to Determine Safety of the DD System
(A) To determine the safety of DD induction in vivo, animals were injected in the striatum with LV expressing MRFP or YFP-DD and LV expressing GDNF-F-DD and MRFP from two separate expression assets (GDNF-F-DD/MRFP). Half of the GDNF-F-DD/MRFP animals were given regular draining water (OFF), and the other half were given TMP (ON). The last group of animals was injected with processed media from cells transduced only with MRFP transfer plasmid (Sham LV). Twelve or 24 weeks after LV delivery, sera were taken for DD/MRFP ELISA, animals were perfused, and their brains were processed for histology. Expression of the system was assayed using immunohistochemistry for DD, GDNF, and MRFP. Toxicity was assessed using immunohistochemistry for NeuN, CD11B, and CD8. (B) Detailed maps of the transfer vectors used for the experiment.
LVs were delivered to the right striatum of SD female rats. The first group (Sham LV) was injected with supernatant from 293T cells transfected with just plasmid expressing monomeric red fluorescent protein (MRFP) (Figure 2B), concentrated as a standard LV, and because it lacked the LV packaging plasmids, contained no functional LV virions. This control group was to account for any potential local inflammation caused by the LV preparation itself. The second group was transduced with LV expressing MRFP and was used as a reporter gene control. The third group of animals was injected with LV expressing DD-regulated yellow fluorescent protein (YFP-DD) (Figure 2B) to determine whether DD could cause inflammation and trigger an immune response. The last group of animals was injected with LV expressing GDNF-F-DD and MRFP (GDNF-F-DD/MRFP). The presence of an independent expression cassette was used to ascertain potential neurotoxicity that could cause loss of transduced cells. Half of the GDNF-F-DD/MRFP group was given 0.2 mg/mL TMP in the drinking water (ON) and the other half was given regular drinking water (OFF).
Twelve or 24 weeks after LV delivery, animals were euthanized, and the brains were processed for histology. To evaluate long-term expression, the brains were stained for DD, GDNF, and MRFP. To determine whether there was a humoral immune response, the sera of animals were collected, and an ELISA against DD and MRFP was performed. To determine whether there was neurotoxicity, the brains were stained for the neuronal marker NeuN. Finally, inflammation was assayed using CD11B staining, and local cellular immune response was assayed using CD8 staining.
Expression and Regulation of GDNF-F-DD in the Brain Can Be Maintained over Long Periods
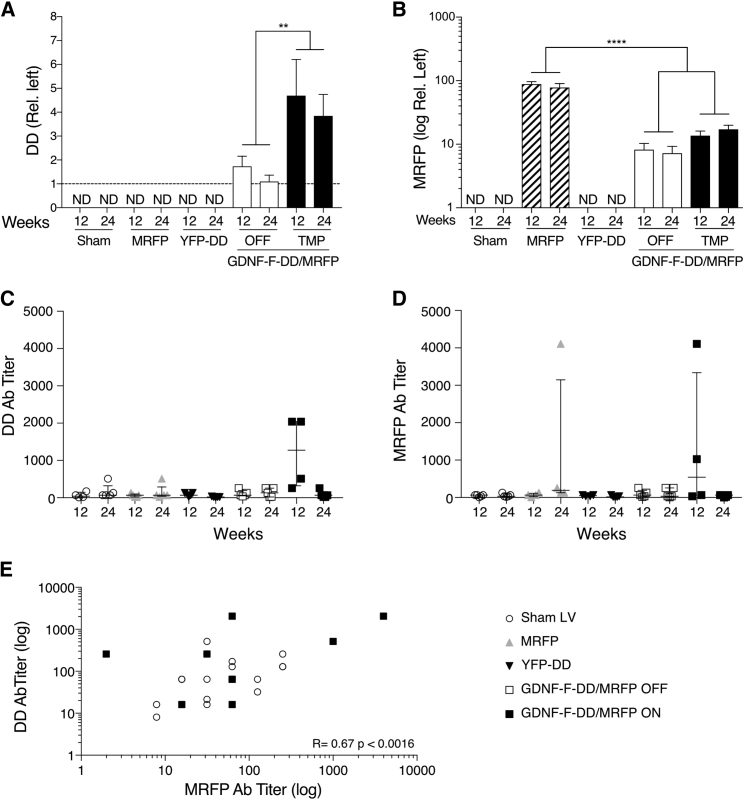
Expression of DD above background could only be detected in the brains from the GDNF-F-DD/MRFP groups (Figures 3A and S2). Therefore, quantification of DD expression by densitometric analysis of coronal sections was performed in the brains from the GDNF-F-DD/MRFP groups, using the contralateral intact side as control. In the GDNF-F-DD/MRFP OFF group, there was minimal DD detected in the intact striatum, 1.7 ± 0.4-fold and 1.1 ± 0.3-fold for 12 and 24 weeks, respectively. Once GDNF-F-DD stabilization was ON, there was an increase in DD accumulation in the GDNF-F-DD/MRFP animals, as the 12-week animals had 4.7 ± 1.5-fold and the 24-week animals had 3.9 ± 0.9-fold DD expression area. A two-way ANOVA comparing the GDNF-F-DD/MRFP groups indicated only significant differences between the TMP and OFF groups, F = 34.3, p < 0.0001; whereas no differences between the two time points were observed.
Figure 3.
DD Regulation Can Be Maintained for Long Periods without Loss of Expression or Transduced Cells, and There Is a Transient Humoral Response against DD and MRFP
(A) Quantification of DD expression was done by densitometry. A two-way ANOVA was performed. (B) Densitometry analysis of MRFP expression was performed in striatal sections. A one-way ANOVA with Tukey multiple comparison tests was performed. (C) An ELISA to detect DD circulating anti-DD antibodies was performed. (D) An MRFP ELISA to detect anti-MRFP antibodies was performed. (E) Correlation between DD and MRFP antibody titers is indicated. n = 4–5 per group.**p ≤ 0.01; ****p ≤ 0.0001. ND, not determined. Error bars represent mean ± SEM or mean + SEM. See also Figure S2.
GDNF expression (Figure S2) was only found in the GDNF-F-DD/MRFP ON and OFF groups and showed GDNF-F-DD regulation consistent with previous experiments. Expression of MRFP (Figures 3B and S2) could only be detected in the MRFP and GDNF-F-DD/MRFP groups. As the MRFP expression in the MRFP group covered the whole striatum, quantification of MRFP signal was performed using densitometry relative to the left untransduced striatum in the MRFP and GDNF-F-DD/MRFP groups. As expected, there was high and consistent expression of MRFP in the 12-week (88.6 ± 8.1-fold) and 24-week (78 ± 11.7-fold) MRFP groups. In the GDNF-F-DD/MRFP groups, there was less expression of MRFP when compared to the MRFP groups. In the GDNF-F-DD/MRFP OFF groups, there was 8.3.6 ± 2.1-fold at 12 weeks and 7.2 ± 2.0-fold at 24 weeks. In the GDNF-F-DD/MRFP ON group, there was 13.7 ± 2.4-fold and 17.2 ± 2.7-fold expression of MRFP in the striatum at 12 and 24 weeks, respectively. A one-way ANOVA of MRFP quantifications indicated differences between groups, F = 32.04, p < 0.0001; and follow-up with Tukey post hoc analysis indicated that there were significant differences in expression between the MRFP groups and GDNF-F-DD/MRFP groups. No differences were detected when the GDNF-F-DD/MRFP groups were compared. The composite expression data of DD, GDNF, and MRFP show that GDNF-F-DD regulation could be maintained in the brain over long periods of time without loss of efficiency.
Simultaneous Expression of MRFP and GDNF-F-DD Resulted in Transient Antibody Responses against DD and MRFP
The sera of the animals were analyzed to determine whether there were circulating antibodies against both DD and MRFP. Briefly, either recombinant DD or MRFP were coated onto 96-well plates, blocked by BSA, and incubated with serial 1:2 (1:2 to 1:4,096) dilutions of the sera from the different animals. The samples were then washed, incubated with anti-rat-HRP (horseradish peroxidase) antibodies, incubated in substrate, and analyzed in a plate reader.
The DD ELISA (Figure 4C) showed that the Sham LV groups had titers of 64.7 ± 37.4 in the 12-week group and 166.4 ± 87.3 in the 24-week group. Similarly, the MRFP group had 57.6 ± 20.6 and 144.0 ± 92.5 in the 12-week group and 24-week group, respectively. As these groups were not exposed to DD, these titer levels were considered background for the assay conditions of the DD ELISA.
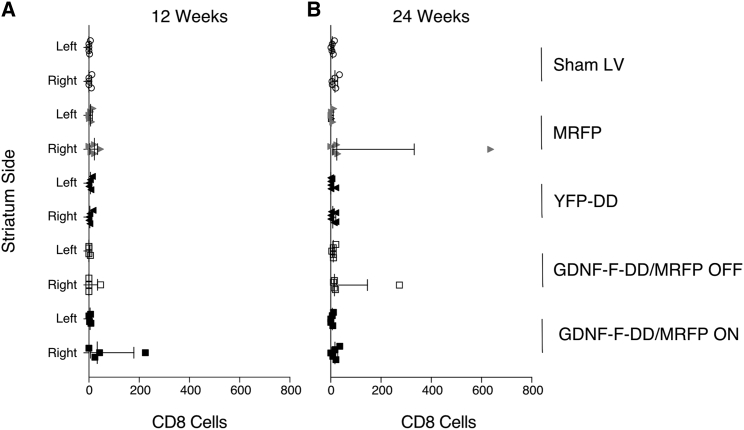
Figure 4.
Mild CD8 Infiltration Observed in Three Animals
(A) Quantification of CD8 cells in sections from animals kept for 12 weeks. (B) Quantification of CD8 cells in striatal sections from animals kept for 24 weeks. n = 4–5 per group. Error bars represent mean + SEM.
The YFP-DD groups exhibited DD titers at background level, with antibodies detected at 80 ± 21.5-fold dilutions for 12 weeks and 16.8 ± 4.4 for 24 weeks. The GDNF-F-DD/MRFP OFF group titers were also at background levels: 106.8 ± 40.0 at 12 weeks and 135.5 ± 53.11 at 24 weeks. However, the GDNF-F-DD/MRFP ON 12-week group had 1,216 ± 483.2. This increase in DD titer was due to 2 animals that had discernible antibody response against DD. Interestingly, no antibody response against DD was present in the 24-week GDNF-F-DD/MRFP ON group, as these animals had titers of 83.2 ± 44.5.
We were also able to analyze 3 animals from the ON-OFF-ON-OFF-ON group in the multiple induction experiment, and in these animals, the DD antibody titer was 53.3 ± 10.7, within the range of control animals.
In the MRFP ELISA (Figure 4D), the Sham LV and YFP-DD groups were used as controls, since these animals were not exposed to MRFP. The Sham LV groups had MRFP titers of 46.4 ± 11.4 at 12 weeks and 54.4 ± 20.0 at 24 weeks. Similarly, the YFP-DD groups had titers of 48 ± 9.2 at 12 weeks and 27.2 ± 10.0 at 24 weeks.
In the MRFP group at 12 weeks, the background titers were 66 ± 24.5. On the other hand, the MRFP titers were 1,152 ± 981.8 in the 24-week MRFP group. This response was caused by one animal that had very high circulating MRFP antibodies. The GDNF-F-DD/MRFP OFF group had 99.2 ± 43.64 at 12 weeks and 116.8 ± 57 at 24 weeks. Similar to the results with the DD ELISA, there was an antibody response against MRFP in the GDNF-F-DD/MRFP ON group at 12 weeks, 1,304 ± 981.8. As with the DD titers, this response was caused by 2 animals that had high MRFP antibody titers. However, this response was not present in the 24-week GDNF-F-DD/MRFP ON group, as these animals had titers of 42 ± 13.6, therefore within background level.
As the 12-week GDNF-F-DD/MRFP ON animals had antibody titers against both DD and MRFP, correlation analysis containing the GDNF-F-DD/MRFP ON and Sham LV groups was performed (Figure 4E). The analysis indicated a significant correlation (Pearson correlation coefficient = 0.67, p = 0.0016) between the MRFP antibody titers and the DD antibody titers in the GDNF-F-DD/MRFP ON groups.
The ELISA data indicate that expression of MRFP alone or induction GDNF-F-DD/MRFP was sufficient to cause a humoral response. There are two interesting points to consider in the humoral response seen in the GDNF-F-DD/MRFP ON 12-week group. First was that the presence of both GDNF-F-DD and MRFP was required to trigger antibody production. Second was that the response was transient and not seen at the 24-week time point. Importantly, the data also show that induction of DD expression alone was not sufficient to cause a humoral immune response.
No Neurotoxicity Observed after Long-Term Regulation of GDNF-F-DD In Vivo
The last step of the study was to verify whether there was a local cellular inflammation or immune response against the DD that might cause neuronal degeneration. To assess direct neurotoxicity, NeuN-positive neurons in the striatum (Figure S3) were quantified with a counting algorithm using the Fiji software suite. To validate the counting algorithm, manually counted sections were compared to automated counted sections using the Fiji software suite. Comparison of the two groups with a paired t test indicated no differences between manual and automated counting (Figure S4).
NeuN automated quantification suggested no difference between the Sham LV group (Table 1) and all other groups either at 12 or 24 weeks. Confirmation using a one-way ANOVA followed by Tukey multiple comparison test did not show any significant differences in NeuN neurons of the different groups.
Table 1.
Lack of Neurotoxicity during Long-Term GDNF-F-DD Expression in the Brain
| No. of Weeks | NeuN Cells (% Left) |
||||
|---|---|---|---|---|---|
| Sham LV | MRFP | YFP-DD | GDNF-F-DD/MRFP |
||
| OFF | ON | ||||
| 12 | 96.6 ± 5.0 | 95.0 ± 6.0 | 94.0 ± 3.6 | 89.0 ± 4.5 | 98.1 ± 2.5 |
| 24 | 99.0 ± 1.0 | 91.7 ± 7.0 | 98.6 ± 2.7 | 99.5 ± 1.6 | 94.0 ± 5.9 |
Quantification is of striatal sections stained for the neuronal marker NeuN.
To determine whether there was a local inflammatory response, the brains were stained for CD11B. As CD11B expression increases during inflammation, densitometric analysis was performed in the brain section, and the left, untransduced side was used as internal control for each section. When compared to the Sham LV, none of the other experimental groups showed any difference in number of CD11B signal (data not shown), and stronger CD11B expression was only observed surrounding the needle tracks (Figure S5). These were the animals with highest CD11B densitometry for each of their respective groups.
Cellular immune reaction was measured by performing CD8 staining. Manual counting was done, as there were very few or no CD8-positive cells per brain in most animals. In the 12-week groups (Figure 4A), Sham LV had 1.8 ± 1.2 CD8 cells in the left striatum and 4.4 ± 2.7 CD8 cells in the right striatum. The MRFP group had 7.4 ± 3.1 CD8 cells in the left striatum and 20 ± 8.0 cells in the right striatum. Similarly, the YFP-DD group had 5.6 ± 2.6 and 5 ± 3.1 CD8 cells in the left and right striata, respectively. In the GDNF-F-DD/MRFP OFF group, only one animal had a detectable number of CD8 cells: 6 in the left hemisphere and 47 in the right hemisphere. In the GDNF-F-DD/MRFP ON group, there were 4.5 ± 1.8 CD8 cells in the left striatum and 73.2 ± 51.7 cells in the right striatum. The high variability in the right hemisphere is due to one animal with CD8 cell infiltration.
In the 24-week group (Figure 4B), there were 7.4 ± 2.4 and 16.6 ± 5.5 CD8 cells in the left and right striata, respectively. In the MRFP group, the left striatum had 5.4 ± 2.0 CD8 cells, and there were 142 ± 124.6 CD8 cells in the right striatum. Once again, the high variability in this group was due to one animal in which infiltration of CD8 cells was discernible. The YFP-DD group had 4.3 ± 3.2 and 9.7 ± 4.0 CD8 cells in the left and right striata, respectively. The GDNF-F-DD/MRFP OFF group had 11 ± 6 CD8 cells in the left striatum and 66.8 ± 52.1 CD8 cells in the right striatum. As in previous groups, this was due to one animal in which mild CD8 infiltration was detectable. In the GDNF-F-DD/MRFP ON group, the left striatum had 5 ± 2.7 CD8 cells and 16.4 ± 6.1 in the right striatum. Overall, the number of CD8 cells varied from animal to animal, and there appeared to be more CD8-positive cells in the injected striatum throughout the different experimental groups.
Interestingly, 3 animals from different groups exhibited CD8 cell infiltration. These animals also had stronger CD11B expression surrounding the needle track and adjacent brain parenchyma. From these 3 animals, only the animal from the MRFP group had a detectable decrease of 12% in NeuN cells. The common factor between these 3 animals was the expression of MRFP. We could not detect any infiltration in the 24-week GDNF-F-DD/MRFP ON group. Moreover, no CD8 infiltration was observed in the YFP-DD group, indicating that DD alone was not sufficient to elicit any long-term cellular immune response.
Discussion
The aim of this study was to fully characterize the potential and safety of the DD system to regulate transgene levels in the brain. For that purpose, GDNF-F-DD was used as a proof-of-principle protein. We have shown previously that the DD system can regulate GDNF expression to therapeutic levels in the brain and that no secretion of GDNF-F-DD or activation of pRPS6 in SNpc was seen when the system was not activated.10
Our present work shows that GDNF-F-DD could be regulated by TMP in a dose-response manner, and at a dosage of 0.2 mg/mL, GDNF-F-DD was induced 20-fold from baseline. There was, however, a significant difference between constitutive GDNF and stabilized GDNF-F-DD levels in vivo. We and others9, 10, 21 have observed such differences when comparing constitutively expressed transgenes and DD-regulated transgenes. This difference seems to be due to TMP dosage, as increasing TMP levels beyond the maximum therapeutic dosage in humans will lead to a proportional increase of DD-regulated transgene to levels similar to that of constitutive transgene expression.9, 10, 21 Therefore, we hypothesize that the dosage of TMP we have used, corresponding to therapeutic TMP levels used in the clinic, is not sufficient to stabilize all the GDNF-F-DD protein being produced, resulting in lower levels of GDNF-F-DD.
We observed that, while the maximum levels of stabilized GDNF-F-DD were roughly half of what we have previously observed in vivo,10 constitutive GDNF levels were much greater that we previously reported. Based on our experience, these differences can be explained by three factors. First, we have observed that, despite similar in vitro functional titers, different viral batches can lead to different levels of in vivo transgene expression (unpublished data). Second, the coordinates and volumes of viral vector injected differ between our previous10 and current experiments. Third, animals used were from a different breeding colony, and the dissections were performed by different personnel. Together, all these factors can lead to the differences between our previous and current results. Importantly, the levels of GDNF activation of SNpc are similar between studies.10
There was a certain degree of variability of pRPS6 cells in SN between individual animals and groups of animals. Although variation of pRPS6 is to be expected, as this protein is dependent on several signaling pathways and is a marker for neuronal activation,22 in our experience, the variability observed was most likely due to antibody batch variation.23 Despite this, our histology and results are in line with published literature using pRPS6 as a marker for activity in the brain.16, 17, 24, 25
Characterization of the kinetics of GDNF-F-DD activation and deactivation indicated that it took up to 4 weeks of GDNF-F-DD to reach maximum levels and 1 week to revert to basal levels. It was also possible to induce GDNF-F-DD stabilization multiple times without loss of activity or expression. Finally, inert GDNF-F-DD regulation could be maintained for over 24 weeks in vivo.
Several patients have developed neutralizing antibodies against recombinant GDNF in clinical trials.26, 27, 28 The humoral response may have been caused by the high amount of GDNF being infused and the bioengineering done to produce the protein in bacteria. Nevertheless, no overt toxicity was observed, and the presence of antibodies did not interfere with the clinical benefit observed.27 Therefore, we decided to focus on the possible immune response caused by the DD component, using MRFP as a reporter gene control. A transient humoral immune response was seen in a few animals of the MRFP and GDNF-F-DD/MRFP ON groups. In these animals, MRFP was expressed alone or in conjunction with GDNF-F-DD. The transient DD antibody response observed in the subset of animals of the GDNF-F-DD/MRFP group was only seen when GDNF-F-DD and MRFP were co-expressed. Moreover, these animals showed a positive correlation between DD and MRFP antibody titers. Several studies in the literature have shown similar humoral immune responses when using LV,29, 30, 31, 32 suggesting that the transient response may have been due to LV virion components, preparation, or vector design.30
Proteasomal degradation of translated proteins is a key step for intracellular antigen presentation to T cells.33 As DD is regulated through proteasomal degradation, we postulated that, if there was a concerted immune response against DD, it would be through a T cell-dependent mechanism in the OFF groups. Histological analysis showed CD8 cell infiltration in the brain parenchyma in a subset of animals. Similar levels of CD8 cell infiltration have been observed when viral vectors are delivered to the brain,31, 32, 34, 35, 36, 37 indicating that the CD8 infiltration was not due to the specific presence of DD.
CD11B expression was elevated surrounding the needle tract in all groups. This is a common finding when injecting viral vectors into the brain and is due to the stereotactic surgery itself.31, 32, 36, 38, 39 In accordance to published studies, the observed local inflammatory and immune responses were asymptomatic, not causing neurotoxicity or loss of transgene expression. Given the variability of immune responses, it could be due to surgical procedures, especially when considering that most of the local immune response and inflammation were contiguous to the injection area.
In addition to the lack of direct neurotoxicity, expression of DD alone neither elicited a humoral response nor caused a significant T cell infiltration or local inflammation beyond the control groups. This is in line with what has been reported in the literature when using viral vector transduction in the brain.31, 36, 37, 38, 39 These data suggest that DD has low immunogenicity when expressed in the brain.
Whenever there is an immune response against the inducible system itself, there is loss of transduced cells with concomitant loss of transgene expression,40 and the inducible system loses its ability to regulate transgene expression.41 No such loss of regulation, transgene levels, transduced cells, or inducibility was observed, as GDNF-F-DD was able to maintain long-term regulation over 24 weeks.
It will be very interesting to determine whether DD can regulate transgene expression in other organs and also whether DD can maintain the same level of safety when regulating transgene expression in more clinically relevant animal models. The preclinical characterization of DD system indicates that it has all the key features8 to potentially regulate gene expression in a clinical setting.
Materials and Methods
Plasmids and Cloning
The plasmid pHR-CMV-GDNF,42 enabling constitutive GDNF expression, and the plasmid expressing GDNF-F-DD alone (pHG-CMV-GDNF-F-DD) have been described in detail previously.10 Gateway technology (Life Technologies Europe, Stockholm, Sweden) was used to clone most of the LVs. To clone the transfer vector expressing MRFP (pHG-EF1a-MRFP), an LR reaction containing an entry plasmid with the human elongation factor 1-alpha (pP4P1-EF1α), p221-MRFP,43 and the destination vector pHG10 was performed according to the manufacturer’s instructions. To clone pHG-CMV-YFP-DD, an LR reaction containing entry plasmid with human cytomegalovirus enhancer/promoter (pP4P1-CMV), entry plasmid with YFP-DD (p221-YFP-DD),9, 10 and the destination vector pHG was performed according to the manufacturer’s instructions. The pCM destination vector was cloned using standard cloning by using the restriction sites Age1 and Kpn1 to insert a DNA fragment containing the human phosphoglycerate kinase promoter driving MRFP expression (PGK-MRFP) into the pHG destination backbone. The PGK-MRFP fragment was amplified by PCR using 10 μM FP-Age1-PGK (5′-CTTATACCGGTCCACGGGGTTGGGGTTG-3′) and 10 μM RP-Kpn1-MRFP (5′-TTCAGGTACCTCAGTACTTGTACAGGGCGC-3′) using the AccuPrime Taq DNA Polymerase System (Life Technologies Europe, Stockholm, Sweden), with pGEM-PGK-MRFP used as a template and according to the manufacturer’s instructions in a 20-μL reaction. The cycling conditions were 95°C for 3 min for initial denaturation, followed by 40 cycles of 95°C for 15 s, 58°C for 30 s, and 68°C for 90 s. The final extension conditions were 68°C for 10 min. The cloning was performed using standard cloning techniques, and the restriction enzymes were purchased from New England Biolabs (New England Biolabs, Ipswich, MA, USA). To clone the lentiviral transfer vector expressing GDNF-F-DD and MRFP (pCM-EF1 α-GDNF-F-DD), an LR reaction containing pP4P1-EF1α, p221-GDNF-F-DD,10 and destination plasmid pCM was performed according to the manufacturer’s instructions.
Cell Culture
Standard cell culture conditions have been described in detail elsewhere.10
LV Production
LV production42, 44 and titration10 have been described in detail elsewhere. The LV used for the experiments had the functional titers of 1 × 109 transducing units per milliliter.
Animals
All animals were housed and handled according to European and Swedish laws. All procedures have been approved and performed according to the guidelines established by the Ethical Committee for Use of Laboratory Animals at Lund University under the permits M09-10 and M366-12. A total of 95 Female SD rats (Charles River Laboratories, Sulzfeld, Germany) weighing 225–250 g were used for the experiments.
Stereotactic Surgeries
The aesthetic solutions and general surgical procedures have been described elsewhere.42 Bilateral injections with triple-injection sites were performed in the following coordinates from bregma. Coordinates for the right hemisphere were: (1) anterioposterior (AP), +1.4 mm; mediolateral (ML), −2.6 mm; and dorsoventral (DV), −5/−4 mm; (2) AP, +0.4 mm; ML, −3.8 mm; and DV, −5/−4 mm; and (3) AP, −0.8 mm; ML, −4.4 mm; and DV −5/−4 mm. Coordinates for the left hemisphere were: (1) AP, +1.4 mm; ML, +2.6 mm; and DV, −5/−4 mm; (2) AP, +0.4 mm; ML, +3.8 mm; and DV, −5/−4 mm; and (3) AP, −0.8 mm; ML, +4.4 mm; and DV, −5/−4 mm. Tooth bar was set at 0, and a total of 6 μL LV suspension (1 μL/DV coordinate) was delivered per hemisphere at the speed of 0.4 μL/min. Unilateral injection coordinates were the same as described earlier for the triple-injection protocol for the right hemisphere.10 A glass capillary was attached to the needle of a 5-μL Hamilton syringe (Hamilton Bonaduz, Bonaduz, Switzerland) to minimize tissue damage in all surgeries.
TMP Treatments
TMP (TMP oral suspension, 10 mg/mL; Meda, Solna, Sweden) was diluted in water to a concentration of 0.05, 0.1, 0.2, or 0.5 mg/mL and given to the animals in their drinking water continuously for the time periods required for the different experiments and groups.
GDNF ELISA
Striatal samples were dissected as described previously10 and diluted in lysis buffer.42 The GDNF ELISA (GDNF Emax ImmunoAssay, Promega Biotech, Nacka, Sweden) was performed according to the manufacturer’s instructions.
Immunohistochemistry
The animals were perfused and processed for histological analysis as described previously.9 Immunohistochemistry was performed as described previously.10 The following primary antibodies were used: rabbit anti-pRPS6 (#2211, 1:400, Cell Signaling Technology, Danvers, MA, USA), goat anti-human GDNF (AF-212-NA, 1:1,000, R&D Systems Europe), rabbit anti-DHFR DD (1:20,000, a kind gift from the Wandless lab that has been used previously21), mouse anti-RFP (ab65856, 1:1,000, Abcam, Cambridge, UK), mouse anti-NeuN (MAB377, 1:1,000, Merck Millipore, Darmstadt, Germany), rat anti-CD11B (MCA275R, 1:1,000, Bio-Rad Laboratories, Solna, Sweden), and mouse anti-CD8 alpha (ab33786, 1:1,000, Abcam, Cambridge, UK). The secondary antibodies used were as follows: biotinylated horse anti-mouse (BA2001, 1:200, Vector Laboratories, Peterborough, UK), biotinylated horse anti-goat (BA9500, 1:200, Vector Labs), and biotinylated goat anti-rabbit-biotin (BA1000, 1:200, Vector Labs).
Quantification of pRPS6 Cells
Three coronal sections were used to quantify pRPS6-positive SNpc neurons: the first coronal section containing the medial lemniscus separating the ventral tegmental area from the SNpc (approximately −5 mm relative to bregma), the adjacent cranial section, and the adjacent caudal section. Data were presented as a percentage of neurons relative to the left intact side.
DD and MRFP ELISA
Sera from animals were harvested at the time of cardial perfusion directly from the heart. Briefly, 1.5 mL blood was harvested with a 17G needle fitted onto a 5-mL syringe and then placed in a microcentrifuge tube. After the blood had coagulated, the samples were centrifuged at 2,000 × g for 10 min. The sera were then carefully pipetted onto a new microcentrifuge tube, aliquoted, and stored at −20°C. DD (endotoxin free, custom-order DHFR-DD, GenScript, Piscataway, NJ, USA) or MRFP (#632503, Clontech Laboratories, Mountain View, CA, USA) was diluted to a concentration of 1 μg/mL in carbonate buffer (0.1 M carbonate buffer; pH 9.6). Then, 50 μL per well of either DD or MRFP solution was added onto a Nunc-Immuno 96-well plate (Thermo Fisher Scientific, Göteborg, Sweden), sealed, and incubated overnight at 4°C. On the following day, the wells were washed 4 times with 200 μL washing buffer (0.05% Tween 20 in Dulbecco’s PBS [DPBS]) and blocked with 200 μL blocking buffer (1% goat serum in DPBS) for 2 hr at room temperature. Afterward, blocking buffer was removed, and 100 μL blocking buffer was added to the second column onward. We added two hundred micrometers of serum from each animal on the first column of each row, and a 1:12 dilution series was performed. The samples were incubated at room temperature for 2 hr. The wells were washed 4 times with 200 μL washing buffer. The last washing buffer was removed, and 100 μL ABTS (Sigma-Aldrich Sweden, Stockholm, Sweden) was added. After 5 min of incubation, the samples were placed in a plate reader, and the absorbance at 650 nm was measured.
Quantification of NeuN and CD8 Cells
Quantification of NeuN and CD8 cell numbers was done using three coronal sections spanning the striatum (approximately 2.28 mm, 1.08 mm, and −0.24 mm relative to bregma). In the first section, three dorsal adjacent fields of the middle portion of the striatum were taken for each hemisphere. In the second and third sections, two adjacent dorsal fields were taken, and two adjacent ventral fields were taken in the middle portion of the striatum. The gray 8-bit images were taken using a 20× objective. The CD8 cells were quantified manually in the Fiji software suite.45 The NeuN cells were batch-quantified in the FIJI software suite using the following Java algorithm: run(“Subtract Background...,” “rolling=50 light sliding”); run(“Normalize Local Contrast,” “blockradiusx=40 blockradiusy=40 standarddeviations=3 center stretch”); setAutoThreshold(“Default”); //run(“Threshold...”); //setThreshold(0, 110); setOption(“BlackBackground,” false); run(“Convert to Mask”); run(“Watershed”); run(“Analyze Particles...,” “size=300-Infinity circularity=0.3-1.00 display exclude clear summarize add”).
Densitometry
Densitometry for GDNF, DD, CD11B, and MRFP signals was measured using three coronal sections spanning the striatum (approximately 2.28 mm, 1.08 mm, and −0.24 mm relative to bregma). The 8-bit gray images were obtained using an image scanner and processed using the Fiji software suite. After inverting the images, pixel densities of GDNF, DD, and MRFP sections were corrected for nonspecific background. Background was not subtracted in CD11B-stained sections, as there are CD11B cells in both sides of the brain. Data for GDNF, DD, and MRFP were expressed as relative of the intact contralateral side. Data for CD11B were expressed as percentage of the intact contralateral side.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 7 (GraphPad Software, La Jolla, CA, USA). Numbers described in the Results section and Table 1 indicate mean ± SEM. Histograms display mean ± SEM. Symbol charts display mean ± SEM. In Figure 2A, the dotted red line was created using non-linear curve fitting. Scatter-dot plots contain individual data points and the median with interquartile range. When a one-way ANOVA was performed, post hoc analysis was done using Dunnett or Tukey multiple comparison tests. Two-way ANOVA was performed in the data displayed in Figure 4B.
Author Contributions
L.Q. and C.L. conceived the study and wrote the manuscript. L.Q., A.N., M.D., L.S.B., P.K., M.A., E.E.-W., and C.I. performed experiments. L.Q., A.N., P.K., M.A., and C.L. analyzed data. L.Q. prepared the figures. C.L. secured funding.
Conflicts of Interest
The authors have no conflict of interest.
Acknowledgments
The authors want to thank Ulla Jarl and Bengt Mattson for expert technical assistance. The authors also want to thank Thomas Wandless for the gift of the anti-DHRF-DD antibody. The work was supported with grants from the Michael J. Fox Foundation, the Swedish Research Council (#2014-3258), the Swedish Brain Foundation, and the Swedish Parkinson Foundation. The work was also supported with infrastructure, personnel, and grants from the BAGADILICO and MULTIPARK consortia at Lund University. The work was performed in Lund, Sweden.
Footnotes
Supplemental Information includes five figures and can be found with this article online at https://doi.org/10.1016/j.omtm.2018.08.008.
Supplemental Information
References
- 1.Naldini L. Gene therapy returns to centre stage. Nature. 2015;526:351–360. doi: 10.1038/nature15818. [DOI] [PubMed] [Google Scholar]
- 2.Senior M. After Glybera’s withdrawal, what’s next for gene therapy? Nat. Biotechnol. 2017;35:491–492. doi: 10.1038/nbt0617-491. [DOI] [PubMed] [Google Scholar]
- 3.Kirik D., Cederfjäll E., Halliday G., Petersén Å. Gene therapy for Parkinson’s disease: Disease modification by GDNF family of ligands. Neurobiol. Dis. 2017;97(Pt B):179–188. doi: 10.1016/j.nbd.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Kordower J.H. AAV2-neurturin for Parkinson’s disease: what lessons have we learned? Methods Mol. Biol. 2016;1382:485–490. doi: 10.1007/978-1-4939-3271-9_32. [DOI] [PubMed] [Google Scholar]
- 5.Olanow C.W., Bartus R.T., Volpicelli-Daley L.A., Kordower J.H. Trophic factors for Parkinson’s disease: to live or let die. Mov. Disord. 2015;30:1715–1724. doi: 10.1002/mds.26426. [DOI] [PubMed] [Google Scholar]
- 6.San Sebastian W., Richardson R.M., Kells A.P., Lamarre C., Bringas J., Pivirotto P., Salegio E.A., Dearmond S.J., Forsayeth J., Bankiewicz K.S. Safety and tolerability of magnetic resonance imaging-guided convection-enhanced delivery of AAV2-hAADC with a novel delivery platform in nonhuman primate striatum. Hum. Gene Ther. 2012;23:210–217. doi: 10.1089/hum.2011.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan K.Y., Jang M.J., Yoo B.B., Greenbaum A., Ravi N., Wu W.L., Sánchez-Guardado L., Lois C., Mazmanian S.K., Deverman B.E., Gradinaru V. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 2017;20:1172–1179. doi: 10.1038/nn.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manfredsson F.P., Bloom D.C., Mandel R.J. Regulated protein expression for in vivo gene therapy for neurological disorders: progress, strategies, and issues. Neurobiol. Dis. 2012;48:212–221. doi: 10.1016/j.nbd.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 9.Tai K., Quintino L., Isaksson C., Gussing F., Lundberg C. Destabilizing domains mediate reversible transgene expression in the brain. PLoS ONE. 2012;7:e46269. doi: 10.1371/journal.pone.0046269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quintino L., Manfré G., Wettergren E.E., Namislo A., Isaksson C., Lundberg C. Functional neuroprotection and efficient regulation of GDNF using destabilizing domains in a rodent model of Parkinson’s disease. Mol. Ther. 2013;21:2169–2180. doi: 10.1038/mt.2013.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwamoto M., Björklund T., Lundberg C., Kirik D., Wandless T.J. A general chemical method to regulate protein stability in the mammalian central nervous system. Chem. Biol. 2010;17:981–988. doi: 10.1016/j.chembiol.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Connor D.M., Boulis N.M. Gene therapy for neurodegenerative diseases. Trends Mol. Med. 2015;21:504–512. doi: 10.1016/j.molmed.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 13.Blits B., Petry H. Perspective on the road toward gene therapy for Parkinson’s disease. Front. Neuroanat. 2017;10:128. doi: 10.3389/fnana.2016.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.d’Anglemont de Tassigny X., Pascual A., López-Barneo J. GDNF-based therapies, GDNF-producing interneurons, and trophic support of the dopaminergic nigrostriatal pathway. Implications for Parkinson’s disease. Front. Neuroanat. 2015;9:10. doi: 10.3389/fnana.2015.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luz M., Mohr E., Fibiger H.C. GDNF-induced cerebellar toxicity: a brief review. Neurotoxicology. 2016;52:46–56. doi: 10.1016/j.neuro.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 16.Decressac M., Kadkhodaei B., Mattsson B., Laguna A., Perlmann T., Björklund A. α-Synuclein-induced down-regulation of Nurr1 disrupts GDNF signaling in nigral dopamine neurons. Sci. Transl. Med. 2012;4:163ra156. doi: 10.1126/scitranslmed.3004676. [DOI] [PubMed] [Google Scholar]
- 17.Chtarto A., Humbert-Claude M., Bockstael O., Das A.T., Boutry S., Breger L.S., Klaver B., Melas C., Barroso-Chinea P., Gonzalez-Hernandez T. A regulatable AAV vector mediating GDNF biological effects at clinically-approved sub-antimicrobial doxycycline doses. Mol. Ther. Methods Clin. Dev. 2016;5:16027. doi: 10.1038/mtm.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gleckman R., Blagg N., Joubert D.W. Trimethoprim: mechanisms of action, antimicrobial activity, bacterial resistance, pharmacokinetics, adverse reactions, and therapeutic indications. Pharmacotherapy. 1981;1:14–20. doi: 10.1002/j.1875-9114.1981.tb03548.x. [DOI] [PubMed] [Google Scholar]
- 19.Le Guiner C., Stieger K., Snyder R.O., Rolling F., Moullier P. Immune responses to gene product of inducible promoters. Curr. Gene Ther. 2007;7:334–346. doi: 10.2174/156652307782151461. [DOI] [PubMed] [Google Scholar]
- 20.Das A.T., Tenenbaum L., Berkhout B. Tet-On systems for doxycycline-inducible gene expression. Curr. Gene Ther. 2016;16:156–167. doi: 10.2174/1566523216666160524144041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cederfjäll E., Broom L., Kirik D. Controlled striatal DOPA production from a gene delivery system in a rodent model of Parkinson’s disease. Mol. Ther. 2015;23:896–906. doi: 10.1038/mt.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biever A., Valjent E., Puighermanal E. Ribosomal protein S6 phosphorylation in the nervous system: from regulation to function. Front. Mol. Neurosci. 2015;8:75. doi: 10.3389/fnmol.2015.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baker M. Reproducibility crisis: blame it on the antibodies. Nature. 2015;521:274–276. doi: 10.1038/521274a. [DOI] [PubMed] [Google Scholar]
- 24.Decressac M., Mattsson B., Weikop P., Lundblad M., Jakobsson J., Björklund A. TFEB-mediated autophagy rescues midbrain dopamine neurons from α-synuclein toxicity. Proc. Natl. Acad. Sci. USA. 2013;110:E1817–E1826. doi: 10.1073/pnas.1305623110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chuang Y.C., Yang J.L., Yang D.I., Lin T.K., Liou C.W., Chen S.D. Roles of Sestrin2 and ribosomal protein S6 in transient global ischemia-induced hippocampal neuronal injury. Int. J. Mol. Sci. 2015;16:26406–26416. doi: 10.3390/ijms161125963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tatarewicz S.M., Wei X., Gupta S., Masterman D., Swanson S.J., Moxness M.S. Development of a maturing T-cell-mediated immune response in patients with idiopathic Parkinson’s disease receiving r-metHuGDNF via continuous intraputaminal infusion. J. Clin. Immunol. 2007;27:620–627. doi: 10.1007/s10875-007-9117-8. [DOI] [PubMed] [Google Scholar]
- 27.Slevin J.T., Gash D.M., Smith C.D., Gerhardt G.A., Kryscio R., Chebrolu H., Walton A., Wagner R., Young A.B. Unilateral intraputamenal glial cell line-derived neurotrophic factor in patients with Parkinson disease: response to 1 year of treatment and 1 year of withdrawal. J. Neurosurg. 2007;106:614–620. doi: 10.3171/jns.2007.106.4.614. [DOI] [PubMed] [Google Scholar]
- 28.Lang A.E., Gill S., Patel N.K., Lozano A., Nutt J.G., Penn R., Brooks D.J., Hotton G., Moro E., Heywood P. Randomized controlled trial of intraputamenal glial cell line-derived neurotrophic factor infusion in Parkinson disease. Ann. Neurol. 2006;59:459–466. doi: 10.1002/ana.20737. [DOI] [PubMed] [Google Scholar]
- 29.Liehl B., Hlavaty J., Moldzio R., Tonar Z., Unger H., Salmons B., Günzburg W.H., Renner M. Simian immunodeficiency virus vector pseudotypes differ in transduction efficiency and target cell specificity in brain. Gene Ther. 2007;14:1330–1343. doi: 10.1038/sj.gt.3302988. [DOI] [PubMed] [Google Scholar]
- 30.Di Natale P., Di Domenico C., Gargiulo N., Castaldo S., Gonzalez Y Reyero E., Mithbaokar P., De Felice M., Follenzi A., Naldini L., Villani G.R. Treatment of the mouse model of mucopolysaccharidosis type IIIB with lentiviral-NAGLU vector. Biochem. J. 2005;388:639–646. doi: 10.1042/BJ20041702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baekelandt V., Claeys A., Eggermont K., Lauwers E., De Strooper B., Nuttin B., Debyser Z. Characterization of lentiviral vector-mediated gene transfer in adult mouse brain. Hum. Gene Ther. 2002;13:841–853. doi: 10.1089/10430340252899019. [DOI] [PubMed] [Google Scholar]
- 32.Abordo-Adesida E., Follenzi A., Barcia C., Sciascia S., Castro M.G., Naldini L., Lowenstein P.R. Stability of lentiviral vector-mediated transgene expression in the brain in the presence of systemic antivector immune responses. Hum. Gene Ther. 2005;16:741–751. doi: 10.1089/hum.2005.16.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blum J.S., Wearsch P.A., Cresswell P. Pathways of antigen processing. Annu. Rev. Immunol. 2013;31:443–473. doi: 10.1146/annurev-immunol-032712-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mazarakis N.D., Azzouz M., Rohll J.B., Ellard F.M., Wilkes F.J., Olsen A.L., Carter E.E., Barber R.D., Baban D.F., Kingsman S.M. Rabies virus glycoprotein pseudotyping of lentiviral vectors enables retrograde axonal transport and access to the nervous system after peripheral delivery. Hum. Mol. Genet. 2001;10:2109–2121. doi: 10.1093/hmg/10.19.2109. [DOI] [PubMed] [Google Scholar]
- 35.Kordower J.H., Emborg M.E., Bloch J., Ma S.Y., Chu Y., Leventhal L., McBride J., Chen E.Y., Palfi S., Roitberg B.Z. Neurodegeneration prevented by lentiviral vector delivery of GDNF in primate models of Parkinson’s disease. Science. 2000;290:767–773. doi: 10.1126/science.290.5492.767. [DOI] [PubMed] [Google Scholar]
- 36.Kordower J.H., Bloch J., Ma S.Y., Chu Y., Palfi S., Roitberg B.Z., Emborg M., Hantraye P., Déglon N., Aebischer P. Lentiviral gene transfer to the nonhuman primate brain. Exp. Neurol. 1999;160:1–16. doi: 10.1006/exnr.1999.7178. [DOI] [PubMed] [Google Scholar]
- 37.Brantly M.L., Chulay J.D., Wang L., Mueller C., Humphries M., Spencer L.T., Rouhani F., Conlon T.J., Calcedo R., Betts M.R. Sustained transgene expression despite T lymphocyte responses in a clinical trial of rAAV1-AAT gene therapy. Proc. Natl. Acad. Sci. USA. 2009;106:16363–16368. doi: 10.1073/pnas.0904514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wong L.F., Goodhead L., Prat C., Mitrophanous K.A., Kingsman S.M., Mazarakis N.D. Lentivirus-mediated gene transfer to the central nervous system: therapeutic and research applications. Hum. Gene Ther. 2006;17:1–9. doi: 10.1089/hum.2006.17.1. [DOI] [PubMed] [Google Scholar]
- 39.Lattanzi A., Neri M., Maderna C., di Girolamo I., Martino S., Orlacchio A., Amendola M., Naldini L., Gritti A. Widespread enzymatic correction of CNS tissues by a single intracerebral injection of therapeutic lentiviral vector in leukodystrophy mouse models. Hum. Mol. Genet. 2010;19:2208–2227. doi: 10.1093/hmg/ddq099. [DOI] [PubMed] [Google Scholar]
- 40.Ginhoux F., Turbant S., Gross D.A., Poupiot J., Marais T., Lone Y., Lemonnier F.A., Firat H., Perez N., Danos O., Davoust J. HLA-A*0201-restricted cytolytic responses to the rtTA transactivator dominant and cryptic epitopes compromise transgene expression induced by the tetracycline on system. Mol. Ther. 2004;10:279–289. doi: 10.1016/j.ymthe.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 41.Latta-Mahieu M., Rolland M., Caillet C., Wang M., Kennel P., Mahfouz I., Loquet I., Dedieu J.F., Mahfoudi A., Trannoy E., Thuillier V. Gene transfer of a chimeric trans-activator is immunogenic and results in short-lived transgene expression. Hum. Gene Ther. 2002;13:1611–1620. doi: 10.1089/10430340260201707. [DOI] [PubMed] [Google Scholar]
- 42.Wettergren E.E., Gussing F., Quintino L., Lundberg C. Novel disease-specific promoters for use in gene therapy for Parkinson’s disease. Neurosci. Lett. 2012;530:29–34. doi: 10.1016/j.neulet.2012.09.059. [DOI] [PubMed] [Google Scholar]
- 43.Campbell R.E., Tour O., Palmer A.E., Steinbach P.A., Baird G.S., Zacharias D.A., Tsien R.Y. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zufferey R., Nagy D., Mandel R.J., Naldini L., Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
- 45.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.