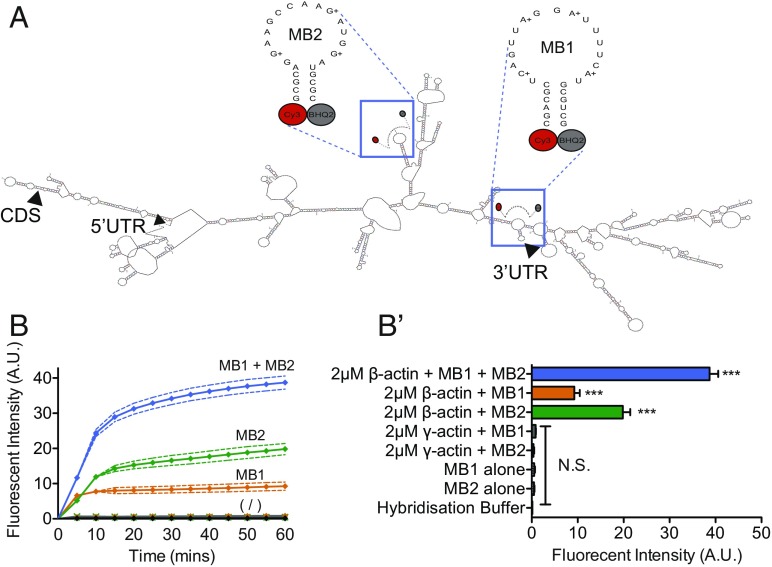
Fig. 1.
Molecular beacons selectively label β-actin mRNA to single-nucleotide precision in vitro. (A) Schematic showing the predicted secondary structure of the sequences of each molecular beacon used to target β-actin mRNA (mFOLD) and the molecular beacon sequences used to target predicted single-stranded regions in the mRNA. Arrowheads indicate the start site of the 5′ UTR, coding sequence (CDS), and 3′ UTR. (B) The addition of 2-μM β-actin mRNA to 1-μM solutions of MB1 or MB2 significantly increased the fluorescent intensity of the solutions compared with MBs in hybridization solution alone and cumulatively amplified fluorescence further when 2-μM β-actin mRNA was added to 1-μM solutions of MB1 and MB2 together (MB1 + MB2). Below the symbol (/) are the changes in fluorescence over time for 1 μM MB1 alone, 1 μM MB2 alone, hybridization buffer alone, 2 μM γ-actin + MB1, and 2 μM γ-actin + MB2. In contrast, the addition of 2-μM γ-actin mRNA did not elicit a change in fluorescent intensity in solutions containing 1 μM of either MB1 or MB2. (B′) For a clearer comparison, B′ shows the fluorescent intensity of each condition at 60 min. ***P < 0.0001 unpaired Students t test; N.S., not significant; n = 3 replicates per condition.