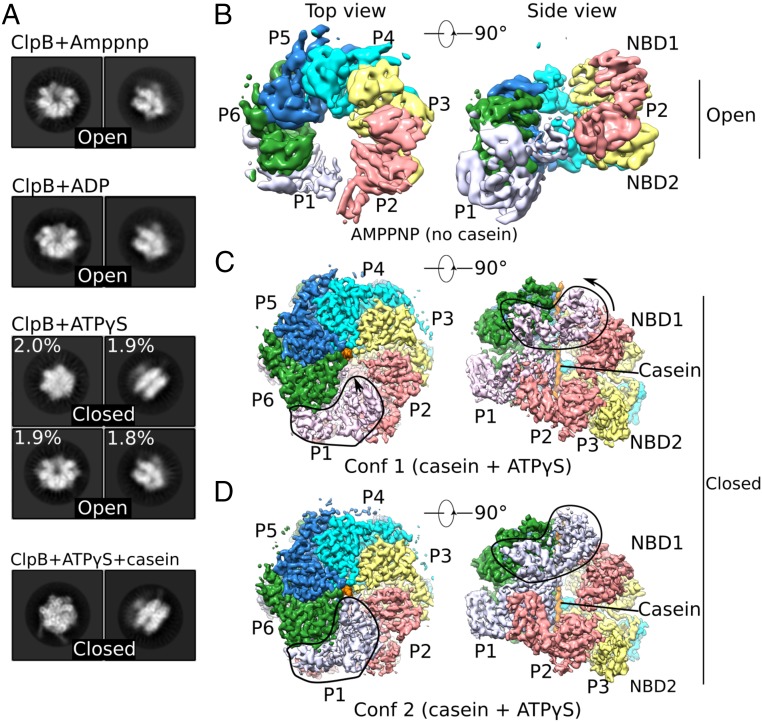
Fig. 1.
Cryo-EM analyses of multiple states of Mtb ClpB disaggregase. (A) Representative 2D images of the ClpB asymmetric hexamer (Left: top view; Right: side view) in the presence of different nucleotides with or without casein substrate. (B–D) Cryo-EM maps of different trapped conformations of ClpB, segmented by individual protomers. B shows an open left-handed spiral structure of ClpB in the presence of AMP-PNP. C and D show two closed conformations of ClpB in a complex with casein and ATPγS. Outlined regions exhibit major conformational differences between these two conformations. Conf 1, conformer 1; Conf2, conformer 2.