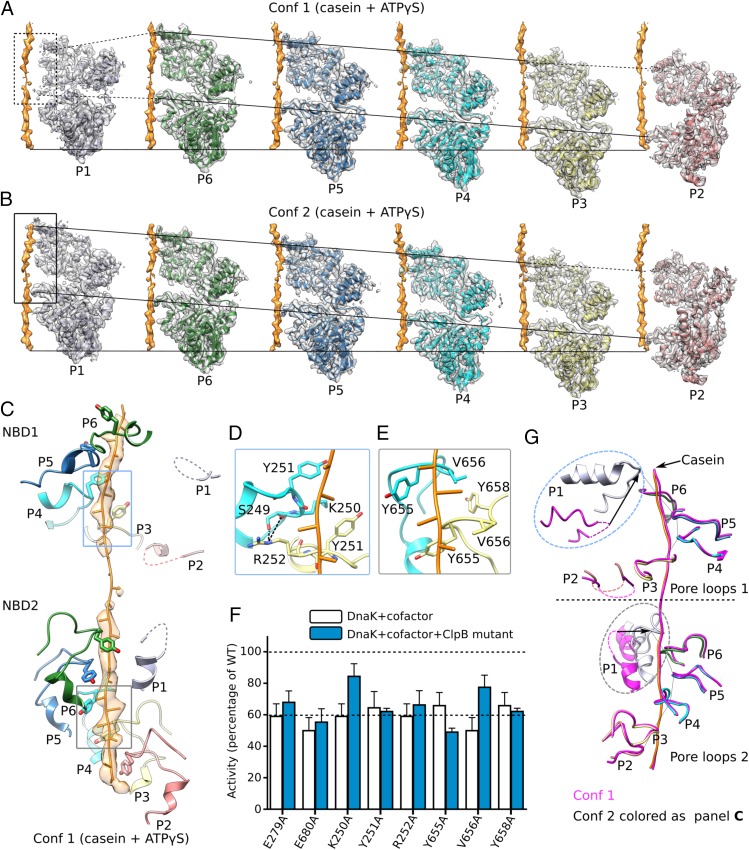
Fig. 3.
Interactions between ClpB and bound substrate mimic casein. (A and B) Individual protomers within conformer 1 (Conf 1) (A) and conformer 2 (Conf 2) (B) are shown separately with a 60° rotation around the bound casein, superposed with their corresponding cryo-EM densities. The casein peptide was aligned across panels (horizontal solid lines) to show the relative height of each protomer in the hexamer. The two slanted lines in each panel connect the pore-loops 1 and 2 across individual protomers, respectively. The dashed lines connect to disordered loops. In both conformations, protomers P3–P6 were fully engaged with the casein through their respective pore-loops 1 and 2, and the tyrosine sidechains of all eight pore-loops had good densities (Fig. 2B). Furthermore, protomer P2 contacted casein via pore-loop 2, but its pore-loop 1 was disordered, so P2 was partially engaged with the peptide. Both pore-loops of P1 were disordered or unengaged in conformer 1 but become fully engaged in conformer 2. (C) In conformer 1, the pore-loops inside the central chamber surrounded and bound the substrate casein in a right-handed spiral mode. This spiral was discontinued at pore-loops 1 of protomers P1 and P2 in the NBD1 ring and at pore-loop 2 of protomer P1 in the NBD2 ring; these pore-loops were flexible and did not contact the substrate. (D) Interactions among neighboring pore-loops 1 and the substrate. Y251 stacked with the casein main chain while R252 H-bonded with S249 of neighboring pore-loops to stabilize the right-handed spiral arrangement. (E) Interactions among neighboring pore-loops 2 and the substrate. Y655 and V656 stacked with the peptide while V656 stacked with Y658 of neighboring loops to stabilize their spiral arrangement. (F) Mutational analysis of the key residues in the pore-loops using a luciferase-based model protein aggregate reactivation assay. The Walker B mutations E279A and E680A were used as negative controls. Data were normalized to reactions containing wild-type ClpB, Hsp20, and DnaK along with cofactors DnaJ1, DnaJ2, and GrpE, which together reactivate denatured luciferase. The dashed line at 60% of wild-type activity defines the yield of reactions lacking ClpB (white bars), indicating the background refolding activity of DnaK and cofactors alone. Hence, reactions with ClpB mutants (blue bars) that show about 60% activity are defined as nonfunctional. (G) A comparison of pore-loops in conformer 1 (magenta) and conformer 2 (colored by protomers as in C). The two conformations were aligned by their respective caseins. Dashed curves mark disordered loops.