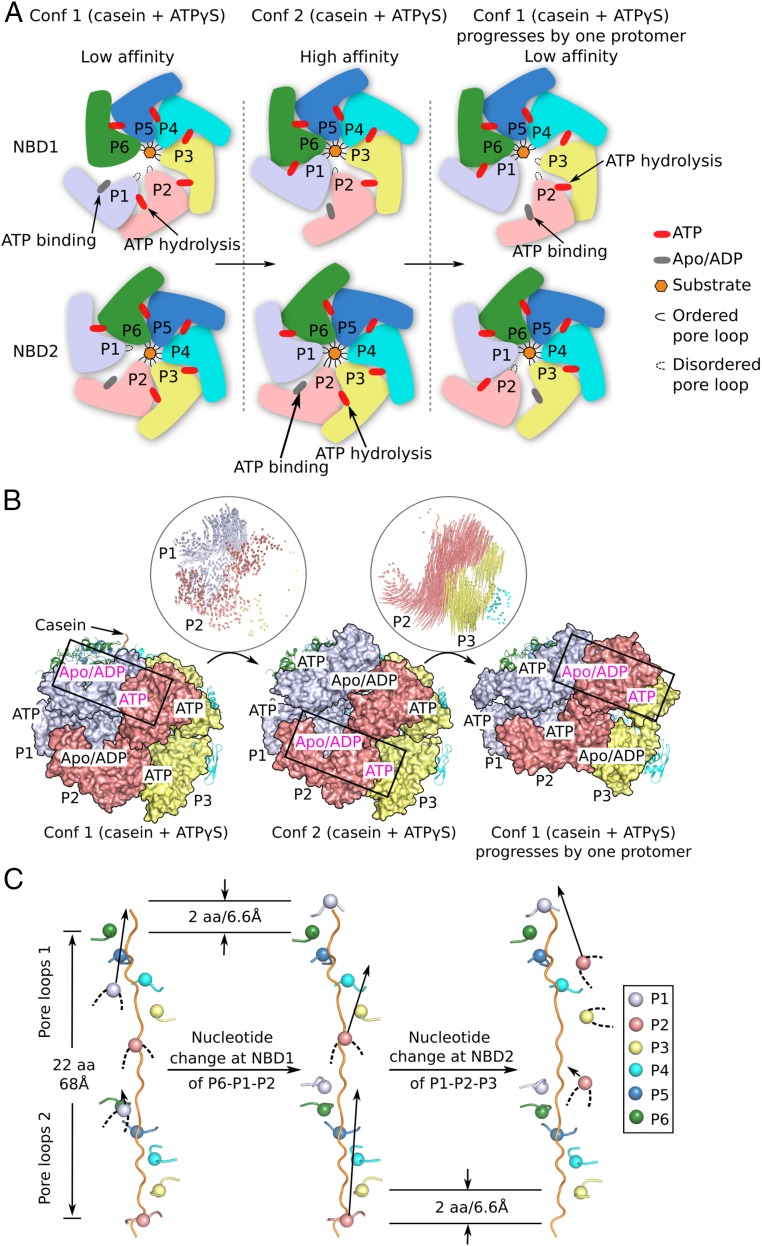
Fig. 6.
A putative three-stage, rotary sequential nucleotide-driven substrate translocation mechanism of Mtb ClpB. (A) A sketch of the proposed mechanism in which the hexamer progresses from stage 1 (conformer 1) to stage 2 (conformer 2) and finally to stage 3. Stage progression is driven by ATP binding and hydrolysis in NBD1 and NBD2 of the two mobile protomers. (B) Atomic trajectories of the progressive conformational changes between stage 1 and 2 and between stage 2 and 3 as illustrated in A. To generate the model for stage 3, conformer 1 was superposed with conformer 2 by aligning protomer P5 in conformer 1 onto protomer P6 in conformer 2. (C) Unidirectional and progressive rearrangements of the 12 pore-loops as the ClpB hexamer goes through the three stages. From stage 1 (Left) to stage 2 (Middle), protomer P1 transits from the unengaged state with none of pore-loops 1 and 2 contacting casein to the fully engaged state with both pore-loops 1 and 2 contacting casein. This is caused by the ATP rebinding to NBD1 of protomer P1 and ATP hydrolysis by NBD1 of protomer P2. From stage 2 (Middle) to stage 3 (Right), protomer P2 transits from a partially engaged state with only pore-loop 2 contacting casein to an unengaged state with neither pore-loop 1 nor 2 contacting casein, and protomer P3 transits from a fully engaged state to a partially engaged state. This step may be caused by ATP rebinding to NBD2 of P2 and ATP hydrolysis by NBD2 of P3. The net effect of going through these three stages is a counterclockwise propagation of conformer 1 by one protomer. Continued cycling through the three stages leads to a rotary sequential model for substrate translocation in Mtb ClpB. Conf 1, conformer 1; Conf 2, conformer 2.