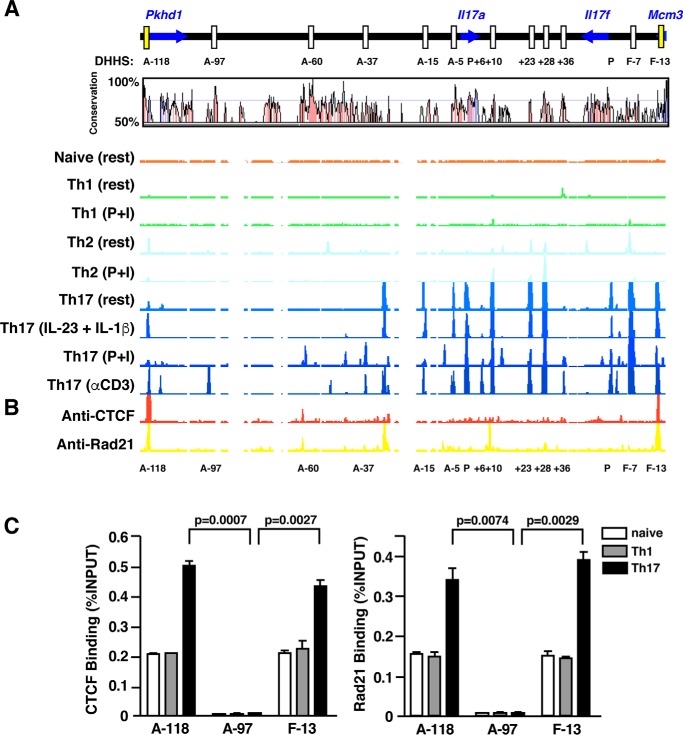
Figure 1.
DNase-hypersensitivity profiles and CTCF/Rad21 binding at the murine Il17a-Il17f loci of naïve, Th1, Th2, and Th17 cells. A, long range DNase hypersensitivity mapping was performed on naïve, Th1, Th2, and Th17 cells. CD4+ T cells isolated from C57/BL6 mice were cultured under Th1 polarizing conditions for 5 days or in Th2 differentiation media for 2 weeks. CD4+ cells isolated from Il17f Thy1.1/Thy1.1 reporter mice were used for Th17 polarization. After 6 days of culture, Thy1.1+ Th17 cells were isolated and rested overnight prior to restimulation. Harvested cells were either left unstimulated (rest), restimulated with PMA + ionomycin (P+I), subject to anti-CD3 treatment (αCD3), or stimulated with rIL-23 + rIL-1β (IL-23 + IL-1β) before DNase I digestion, labeling, and hybridization to custom arrays for DNase-chip analysis. DNase I hypersensitivity profiles are displayed using the IGB browser (Affymetrix) and are aligned with a corresponding VISTA plot demonstrating percentage similarity between mouse and human sequences. CNSs are defined as noncoding regions of at least 100 bp in length exhibiting greater than 70% sequence homology between species. Positions of CNSs are relative to the transcription start of the mouse Il17a or Il17f gene. B, ChIP-chip analysis of CTCF and Rad21/cohesin occupancy across the extended Il17a–Il17f loci. Thy1.1+ cells generated from Il17f Thy1.1/Thy1.1 reporter mice underwent chromatin immunoprecipitation (ChIP) with an antibody directed against murine CTCF or Rad21. ChIP samples and input material were subject to whole genome amplification and hybridized to custom-tiled arrays. Regions of CTCF and Rad21 binding are visualized using the IGB browser (Affymetrix). C, CTCF and Rad21ChIP analysis in naïve, Th1, and Thy1.1+ Th17 cells. Results are the mean ± S.E. of duplicate determinations and are representative of two independent experiments; results were quantified using real-time PCR, and data are expressed as percentage of input DNA recovered.