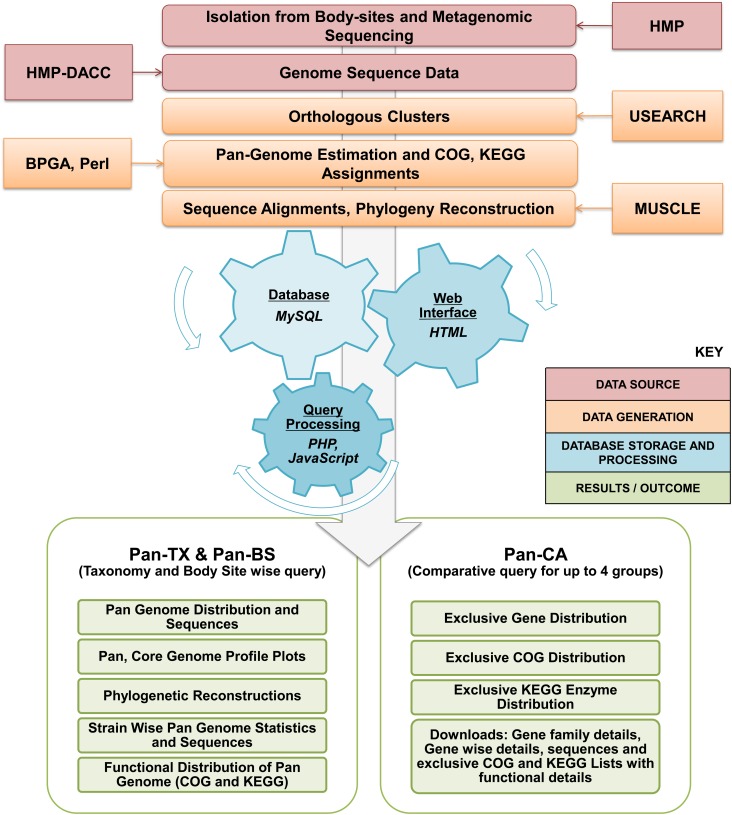
FIGURE 1.
Workflow for PanGFR-HM. The workflow involves preprocessing of the whole genome records from GenBank. The protein sequences from all the available strains are then clustered using USEARCH at various sequence identity cut off levels. The gene presence/absence is determined and stored as a binary matrix using BPGA Pipeline. The tabular data generated after sequence processing and functional assignments are then uploaded into separate MySQL databases for respective sequence identity cut offs. Majority of the backend programs are written in PHP. Finally, various pan genomic and comparative analysis results generated from the set of strains and clustering parameters selected by the users are then provided via the web browser.