Abstract
The complex karyotype (CK) is an established negative prognostic marker in a number of haematological malignancies. After the introduction of effective mitogens, a growing body of evidence has suggested that the presence of 3 or more aberrations by conventional banding analysis (CBA) is associated with an unfavorable outcome in chronic lymphocytic leukemia (CLL). Thus, the importance of CBA was recognized by the 2018 guidelines of the International Workshop on CLL, which proposed the introduction of CBA in clinical trials to validate the value of karyotype aberrations.
Indeed, a number of observational studies showed that cytogenetic aberrations and, particularly, the CK may have a negative independent impact on objective outcome measures (i.e. time to first treatment, progression free survival, time to chemorefractoriness and overall survival) both in patients treated with chemoimmunotherapy and, possibly, in patients receiving novel mechanism-based treatment.
Here, we set out to present the scientific evidence supporting the significance of CK as a prognostic marker in CLL and to discuss the biological basis showing that the CK is a consequence of genomic instability.
Keywords: chronic lymphocytic leukemia, complex karyotype, prognosis, target therapy, Richter transformation
INTRODUCTION
Chronic lymphocytic leukemia (CLL) is the most common leukemia of the adult in western countries [1–2]. As a result of genetic and biological complexity [3], the disease runs a variable clinical course, with some patients showing a very indolent evolution and others rapid progression [4]. Several biological features were identified which may predict the time between the diagnosis and the disease progression, the overall survival (OS) (i.e. prognostic markers) and the quality of response to a specific agent (i.e. predictive markers) [5]. The mutational status of the variable portion of the immunoglobulin gene (IGHV) [6], chromosome 17p deletion as detected by fluorescence in situ hybridization (FISH) [7] and TP53 gene mutations [8] represent powerful prognostic/predictive factors commonly used to stratify CLL patients into different risk groups in the era of chemoimmunotherapy [5, 9–10]. The introduction of effective mechanism-based treatment (i.e. ibrutinib, idelalisib, venetoclax) significantly improved the outcome of CLL [11–13] and the adverse impact of 17p-/TP53 mutation and of the IGHV mutational status were found to be attenuated in several studies [14–16].
Cytogenetic abnormalities in CLL were first described using chromosome banding analysis (CBA) in the 1970s [17] and in 1990 the complex karyotype (CK) was found to be associated with shorter survival [18]. At that time conventional cytogenetics was limited by the low number of dividing cells in culture [19]. Since 2006, the introduction of the effective mitogens CpG-oligodeoxynucleotide (DSP30) and Interleukin (IL)-2, dramatically improved the mitotic yield [20–21] and 65-83% of CLL patients were shown to carry a karyotype aberration [22–24]. Furthermore, evidence was provided that 21.5-35.7% of CLL cases without aberration by FISH carried chromosome aberrations by CBA, including CK, that were associated with an inferior outcome [25]. Overall, a CK is detectable in 14-34% of untreated CLL patients [23–24, 26–29], and up to 25-35% in the relapse refractory setting [30].
The CK has a strong adverse prognostic significance in several hemopoietic neoplasms such as acute myeloid leukemias, myelodysplastic syndromes and myelofibrosis [31–36] and a number of papers were published in the last 10 years highlighting its relevance in the landscape of prognostic biomarkers in CLL. We therefore set out to review here the biological basis underlying the development of CK and the prognostic and predictive value of this cytogenetic pattern in CLL in the chemoimmunotherapy and the mechanism-based treatment era.
METHODS
In this analysis the CK was defined by the presence of at least three clonal aberrations in the same clone as detected by CBA [18, 37–39]. The studies describing the significance of multiple unrelated clones with less than 3 chromosome aberrations in the same clone were excluded.
Literature search
To describe the biological role of CK and its association with other biomarkers in CLL we performed a first search on PubMed using a MeSH controlled vocabulary using the following terms: “Leukemia, Lymphocytic, Chronic, B-Cell” [Mesh] AND “Karyotype” [Mesh] OR “Abnormal Karyotype” [Mesh] OR “Karyotyping” [Mesh]. We found 311 citations without any restriction on publication date. We included in this analysis those paper fulfilling the following requirements: i) English language; ii) biologic characterization including salient clinicobiologic parameters and CBA; iii) single centre or multicentre studies enrolling consecutive patients and studies using a learning cohort and a validation cohort.
We also performed a research on PubMed to identify publications from January 2000 to March 2018 describing the role of the CK as a prognosticator in CLL patients (chemoimmunotherapy and targeted therapy era). The following terms were used: “Leukemia, Lymphocytic, Chronic, B-Cell/drug therapy”[Mesh] AND “Clinical Trial” [Publication Type]. We included in this review only full length manuscripts satisfying these criteria: i) English language; ii) phase 2 or phase 3 clinical trials; iii) multivariate and/or univariate analysis including CK; iv) time to first treatment (TFT), or progression free survival (PFS) or overall survival (OS) as clinical endpoints. Manuscripts describing the prognostic impact of the selected parameters in patients who had received experimental treatment were not included.
RESULTS
CK as a consequence of genomic instability
Considering that up to 90% of CLL with CK show an U-IGHV mutational status [22–23, 28–29] a relationship may exist between the IGHV gene configuration and the development of CK. Indeed a large body of evidence showed that the lymphocytes with U-IGHV i) respond to antigen stimulation by activating intracellular signalling, ii) undergo cell divisions in vivo as shown by incorporation of deuterated water, iii) carry relatively shorter telomeres and, iv) tend accumulate genomic defects [40–41]. Interestingly, Burns and co-workers [42], used a whole exome sequencing approach to study gene mutations in correlation with the IGHV gene configuration and found that exonic CLL driver gene lesions were more common in U-IGHV CLL than in CLL with mutated IGHV gene. Coding mutations involved NOTCH1, SF3B1, TP53, KLHL6 and, less frequently, IKZF3, SAMHD1 and BIRC3[42]. These gene mutations may directly increase genome instability reducing the ability of the cells to respond to DNA damage and may also act in an indirect manner, affecting pathways linked to cell proliferation or serving as an important bridge with the microenvironment, which is of particular importance in CLL [43].
Thomay et al[44], reported that loss or mutation of TP53 was associated with an increased number of break events, with frequent involvement of (near-) heterochromatic regions adjacent to the centromeres, generating dicentric chromosomes and whole-arm translocations. In a recent analysis on relapsed/refractory (R/R) CLL, TP53 mutations preceded clonal evolution leading to the emergence of clones with CK [45]. Furthermore, patients with TP53 mutations showed significantly shorter telomeres [44, 46–48] a condition causing chromosomal instability [44, 49]. Though few data are available on the association between CK and telomere length, it is worth noting that two studies showed that patients with CK had shorter median telomere length [44, 50]. In patients with 11q-/ATM deletions and a CK, the frequency of TP53 mutations was significantly lower than in patients with CK without del (11q), suggesting that the disruption of the DNA damage control pathway through ATM or TP53 lesions may favour the development of multiple chromosomal rearrangements [44]. Other mutations occurring at a higher incidence in patients with CK involved FBXW7 (16.7%) in a study [24] and MYD88 (14.3%) in another study [26]. These genes have been linked to the NOTCH1/WNT pathways and to the inflammatory pathway, respectively [51]. FBXW7 encodes for a tumour suppressive protein, which regulates ubiquitin-mediated degradation of various oncoproteins (cyclin E, c-MYC, NOTCH) [52]. The abnormal binding of cyclin E to FBXW7 has been related to chromosomal instability in hematopoietic progenitors [53] providing a possible functional link to the development of CK. MYD88 mutation may have a role in generating genome instability through the activation of the RAS/ERK pathway [54]. Moreover, a recent study by Oliveira-Santos et al pointed out a possible role of the histone methyltransferases SET and MYND domain containing 2 (SMYD2) and SET and MYND domain containing 3 (SMYD3), members of the SMYD family of methyltransferases, in the development of CK [55]. In this study, SMYD2 and SMYD3 were found to be overexpressed in CLL patients. Interestingly, lower expression of SMYD2 and SMYD3 was significantly associated with a CK [55]. Although the mechanism linking these methyltransferases and CK is unknown, it noteworthy that SMYD2 may act as an oncogene by promoting the methylation of p53 and of the retinoblastoma tumor suppressor protein (RB) [56–57], and that SMYD3 promotes MAP3K2 methylation, inducing genomic instability by activation of Ras/Aurora kinase A-driven mechanisms [55, 58–59].
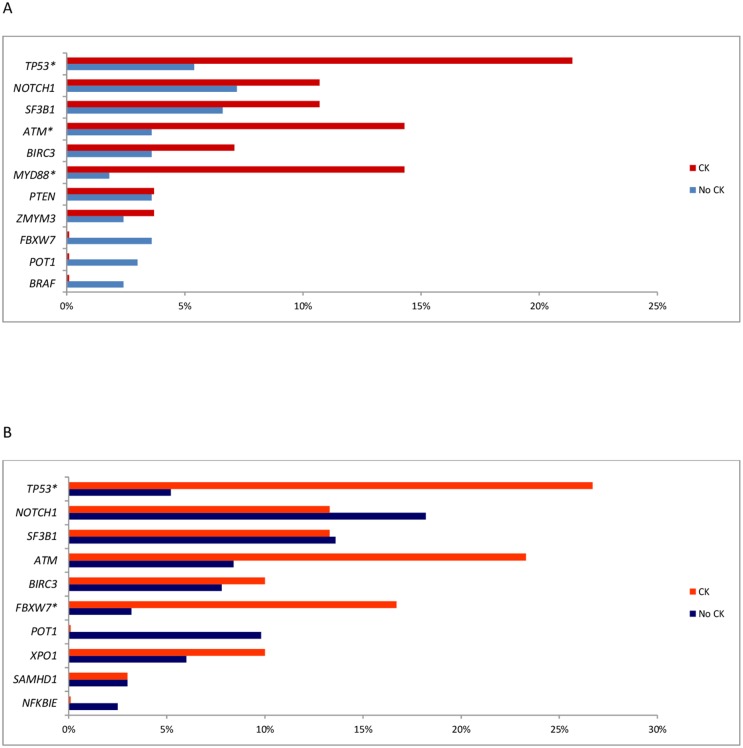
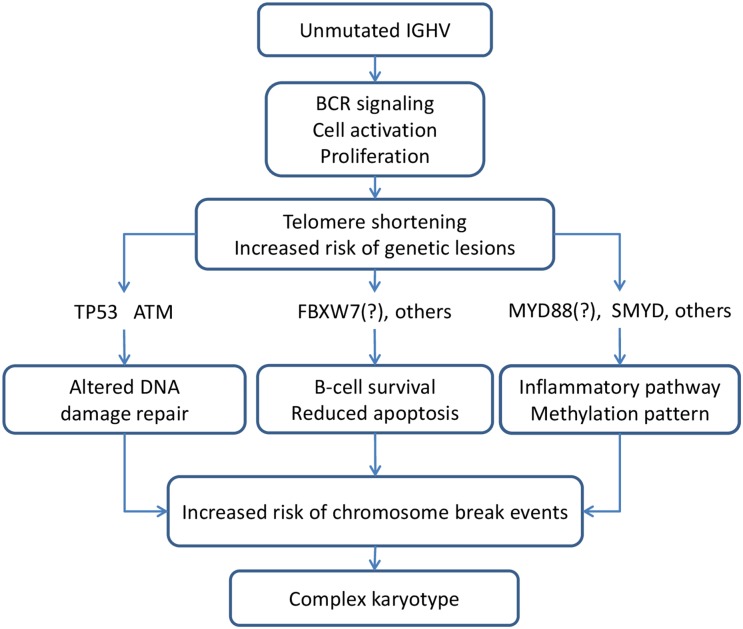
Although the precise mechanisms underlying the development of CK in CLL are have not been elucidated, evidence was provided in 2 studies using CBA and NGS on a panel of CLL driver genes that the CK may be associated with a distinct pattern of genetic lesions (Figure 1). A sequence of events possibly leading to the development of complex cytogenetic rearrangements is illustrated in Figure 2.
Figure 1.
(A) Frequency of Gene Mutation by NGS in patient with or without CK reported by Rigolin et al [26] (B) Frequency of Gene Mutation by NGS in patient with or without CK reported by Herling et al [24]. *p<0.05
Figure 2. Genetic background favouring the development of complex karyotype.
The CK is more frequently found in CLL with unfavorable clinicobiologic features
An extensive biologic characterization of CLL patients enrolled in clinical trials allowed for the identification of biomarkers associated with an inferior prognosis in large international studies recruiting patients treated with chemoimmunotherapy [10, 60]. Because CBA was not included in these studies, we set out analyse possible correlations between CK and clinical parameters (stage, age, sex, performance status) and biomarkers, i.e TP53 disruption, 11q-; unmutated IGHV gene (U-IGHV) with a documented prognostic significance at multivariate analysis in the chemoimmunotherapy era [10, 60].
Studies including CBA and the classical clinicobiologic prognosticators showed that no consistent correlation was found between CK and unfavorable clinical parameters, such as age, sex, performance status, beta-2-microglobulin levels, whereas an association appears to be well documented between CK and advanced stage, as shown in Supplementary Table 1.
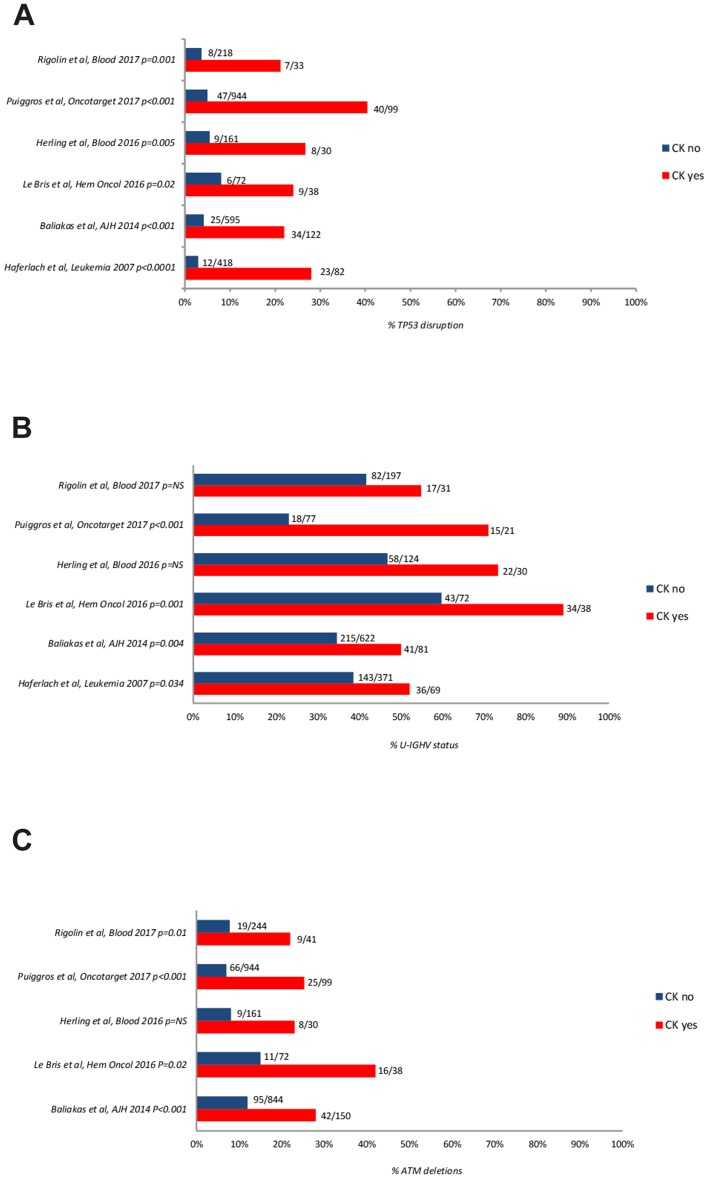
The U-IGHV status, del (17p)/TP53 mutations or del (11q)/ATM deletions were more frequently seen in CLL with CK, than in CL without CK (Figure 3) [22–24, 28–29, 39, 61–63].
Figure 3.
Frequency of TP53 disruption (A), ATM deletion (B), and unmutated IGHV gene configuration (C) according to the presence or absence of CK. The number of cases are reported aside each bar. NS: not significant; UM-IGHV: unmutated IGHV.
A number of recurrent driver gene mutations was detected in CLL by next generation sequencing (NGS) and excellent reviews highlighted that, while the vast majority of them recur across patients at a low frequency, mutations of TP53, ATM, NOTCH1, and SF3B1 can be found in >5% of treatment-naïve patients and may predict for an inferior prognosis [48, 64–65]. We therefore analysed available evidence on the association between genetic features and CK. Several studies reported a correlation between CK and TP53 or ATM mutations or the unmutated IGHV configuration (Figure 3). The results of 2 studies using CBA with novel mitogens and NGS to detect mutations in large CLL-specific gene panels are shown in Figure 2. Overall, these data show that the incidence of TP53 disruption in CLL with CK was significantly higher (21-80% of the cases) than in an unselected treatment-naïve CLL population (3-13% of the cases) [22–24, 26, 28–29, 63] or in CLL without CK. Likewise, a statistically significant association was found between CK and the unmutated IGHV (U-IGHV) in the majority of studies (50-90% of the cases) [22–24, 28–29, 63] and with 11q- or ATM mutation (22%-42, 1%) (Supplementary Table 1, Figure 1) [23, 26–27, 29, 63]. Herling and colleagues did not find a significant association between CK and the U-IGHV status in the patients enrolled in the CLL11 trial [24], possibly due to over-representation of IGHV-unmutated cases and consequent low number of IGHV-mutated cases in this analysis including patients with disease progression. These findings are not surprising, since TP53 and ATM function are involved in maintaining genomic stability [66–67], and the U-IGHV configuration identifies a CLL clone that is responsive to B-cell receptor antigen stimulation with consequent cell activation [40–41].
We also analysed possible association of CK and immunophenotypic markers. In 3 studies the CK was associated with CD38-positivity [23, 63] (Supplementary Table 1). No study analysed whether a CK was associated with CD49d expression, an immunophenotypic marker of unfavorable prognosis [68].
Overall the data here summarized clearly show that there is a strong and reproducible association of CK with advanced stage, U-IGHV, TP53 disruption, ATM mutations.
The CK is a strong adverse risk factor in the chemoimmunotherapy era
The CK was shown to represent an independent adverse prognostic factor in several studies analysing a full set of biomarkers and using robust efficacy endpoints, i.e. time to first treatment (TFT), progression free survival (PFS) or overall survival (OS).
Impact on TFT
Early observations concerning the negative impact of complex chromosomal abnormalities on clinical outcome date back to the first studies on cytogenetic aberrations in CLL [18–19, 69–71]. In a monocentric study on 109 CLL patients conducted by Mayr et al, the CK was found to be predictive of a worse TFT at univariate analysis (median TFT, 26 months; 95% CI, 15-37 months vs 106 months 95% CI, 61-151 months; P < 0.001) [20]. In another monocentric study on 482 CLL patients [38], the CK was associated with a shorter TFT in treatment-naïve patients at univariate analysis (HR 1.644; P = 0.029) and similar findings were reported by Travella and coworkers, who observed a 13-month median TFT in patients with CK as compared with 69-month median TFT in patients without CK (P=0.015) [72]. These observations were also reported in a larger study on 1001 previously untreated CLL where the CK was associated with a shorter TFT at univariate (p=0.01) and multivariate analysis [23]. In subsequent studies the prognostic impact of CK on TFT was documented in a learning cohort (LC) including 166 patients and in an independent validation cohort (VC) including 250 patients (HR 5.004; 95% CI, 1.980-12.597; P<0.001 and HR 2.159; 95% CI, 1.499-5.331; P<0.001, respectively). The difference held at multivariate analysis in the LC (HR 2.418; 95% CI; 1.173–4.983; P=0.017) [39]. More recently, Puiggros et al, conducted a study on 1045 untreated MBL/CLL patients to evaluate the clinical impact of CK and high risk FISH (HR-FISH) on outcome [63]. The cases with CK had a higher 2-year cumulative incidence of progression requiring treatment (48%; 95% CI, 36-58% vs. 20%; 95%CI, 18-23%; P<0.001) [63]. Finally, in a large retrospective monocentric study by Rigolin et al including 335 newly diagnosed CLL, the CK was associated with a shorter TFT, independent from CLL-IPI (HR 2.157; IC 95%, 1.177-3.952; 0.013) [27].
Impact on PFS
The correlation between CK and PFS was investigated in few clinical trials in the era of chemoimmunotherapy (CIT). In a study by Badoux et al, the CK proved to be predictive of shorter PFS in 284 R/R CLL patients treated with FCR (HR 2.6; 95%CI, 1.5-4.4; P<0.001) [73]. Similar data were presented in another study including 80 R/R CLL patients treated with Cyclophosphamide, Fludarabine, Alemtuzumab, and Rituximab [74]. In this analysis the CK along with del (17p) were significantly associated with worse PFS (HR 4.1; 95%CI, 2.0-8.4; P<0.001). Likewise a retrospective study on 110 CLL patients treated with first-line FCR showed that the CK was associated with shorter median PFS (21 vs. 55 months; HR 2.4; 95%CI, 1.14-5.19; P=0.002) [29]. Herling and coworkers described the impact on outcome of CK in a subset of patients treated with chlorambucil-based chemo- or chemoimmunotherapy in the CLL11 trial and found that chromosome translocations, a type of chromosome imbalance often associated with complex karyotype [75], was associated with shorter PFS [24].
Impact on OS
As shown in Table 1, a significantly shorter survival was observed in virtually all the observational studies that included treatment naïve patients, including a monocentric study on CLL patients treated with first-line FCR [29] and an analysis using a learning cohort and a validation cohort [39]. Interestingly, the presence of CK was shown to represent an adverse prognostic parameter that was independent of the international prognostic index [27].
Table 1. Impact of complex karyotype on OS.
| Tot. pts | CK | Treatment | Disease status | Median OS (months) |
Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Reference | CK yes | CK no | HR (IC 95%) | P | HR (IC 95%) | P | ||||
| [20] | 109 | NA | various | TN and pretreated | 107 | 346 | 15.44* | <0.001 | - | Ø |
| [38] | 482 | 71/399 (17.8%) |
NA | TN | 81.1% at 5y | 86-94.4% at 5y # |
3.830 (1.714-8.560) |
0.001 | - | Ø |
| [73] | 284 | 22/182 (12.1%) |
FCR | R/R | 26 | 10.5 m. -78 m. ## |
- | - | 1.9 (1.1-3.2) |
0.015 |
| [74] | 80 | 8/67 (11.9%) |
CFAR | R/R | NA | NA | - | - | 2.0 (1.1-3.7)** |
0.022 |
| [72] | 38 | 16/38 (42.1%) |
NA | TN | 56 | 144 | - | - | - | - |
| [39] | LC 166 VC 250 |
20/145 (13.8%) 35/238 (14.7%) |
various | TN | NA | NA | 2.701 (1.988-8.787) 2.155 (1.160-4.004) |
<0.001 0.015 |
4.856 (1.475-9.998) 3.630 (1.358–9.703) |
<0.0001 0.010 |
| [29] | 110 | 38/110 (34.5%) |
FCR | TN | 72.4% at 5 y | 85.8% at 5y | - | - | 5.16 (1.2-22.1) |
0.07 |
| [24] | 161 | 30/154 (19.5%) |
Clb/Clb-R/Clb-G | TN | NA | NA | 2.9 (1.5-5.4)° 2.6 (1.3-5.4)°° |
0.001° 0.006°° |
2.682 (1.366-5.264) |
0.004 |
| [63]# | 1045 | 99/1043 (9.5%) |
various | TN | 79 | NR | - | - | 1.66 (1.06-2.59) |
0.027 |
| [27] | 335 | 41/287 (14.3%) |
various | TN | 70 | 135 | 3.176 (1.882-5.359) | <0.001 | 3.572 (1.341-9.515) |
0.011 |
| [77] | 186 | 37/186 (19.8%) |
Lenalidomide-R Lenalidomide-O |
R/R | 23 | 62.8 | - | - | 2.08 (1.15-3.76) |
0.015 |
TN=treatment naïve; R/R= relapsed/refractory; OS=overall survival; CK=complex karyotype; NA=not available; Clb=chlorambucil; FCR=Fludarabine, Cyclophosphamide, Rituximab; G=Obinutuzumab; R= Rituximab; O-Ofatumumab; Ø=not significant; LC: learning cohort; VC: validation cohort
*log rank; **CK or 17p aberration; °all arms; °°(Clb-R+Clb-G);
#MBL/CLL
# OS was compared in this study between patients with CK and patients without CK carrying chromosome translocations, or 1-2 aberrations or normal karyotype.
## OS in the patients without CK was reported in this study according to the aberration detected by FISH (13q-, +12, 11q-, 17p).
In 2016, Herling et al[24], performed an analysis in a subgroup of 161 elderly and comorbid patients enrolled in the CLL11 trial comparing the efficacy of chlorambucil alone, against chlorambucil with rituximab or with the second generation anti CD20 obinutuzumab. The patients with available karyotype included in analysis were representative of the entire study cohort with the exception of a higher percentage of patients >70 years. A CK in 30/154 (19.5%) patients was shown to represent an independent negative prognostic factor on OS along with advanced stage, elevated beta-2-microglobulin (β2M) an unmutated IGHV gene and mutations in the POT1 gene. Interestingly, in a phase II trial evaluating chlorambucil and rituximab in 85 treatment naïve patients [76] the presence of CK was associated with a lower overall response rate (ORR) and complete remission rate, which represent surrogate endpoints of OS. Indeed Takahashi et al, more recently confirmed the adverse impact of CK on ORR and OS in a study conducted on 186 R/R CLL patients treated with a Lenalidomide based regimen [77]. Finally, in two trials including relapsed/refractory CLL, the CK was significantly associated with a shorter OS in patients treated with fludarabine cyclophosphamide and rituximab, with or without alemtuzumab [73–74].
CK is an independent adverse prognosticator in high risk CLL
A negative prognostic impact of CK was recently documented even when the analysis was restricted to high risk CLL as defined by unfavorable genetic features [23–24, 28, 78]. Herling et al, showed that patients with CK alone exhibited a similar survival as those with TP53 lesion, whereas patients with both CK and TP53 aberrations had a particularly poor prognosis (P<0.001) [24]. In another study on 101 patients carrying TP53 abnormalities, 31/101 cases (47%) showed a CK. CK was associated with shorter OS at multivariate analysis (HR 2.47; 95% CI, 1.11–5.49; P=0.027) confirming that CK may portend a dismal outcome in this aggressive subgroup [78].
In a study on previously untreated high-risk CLL (defined by the presence of U-IGHV gene and/or chromosome 11q22 deletion and/or chromosome 17p13 deletion and/or TP53 mutations) the presence of CK in 21/101 (20.8%) cases has been associated with unfavorable FISH (i.e. 11q- or 17p-) (P<0.001) and TP53 disruption (P=0.012). In multivariate analysis, the CK was significantly associated with a shorter TFT (HR 2.934; 95% CI, 1.686-5.108; P<0.001), an inferior OS (HR 2.914; 95% CI, 1.080-7.861; P = 0.024) and a trend toward a shorter time to chemorefractoriness (TTCR) (HR 2.486; 95% CI, 0.905-6.825; P=0.077) [28].
In the subgroup of patients with concurrent TP53 disruption and unfavorable karyotype, the median TFT reported in this study was 5.5 months compared to 97.3 months in patients with sole TP53 disruption (p<0.0001). Likewise, the median OS and TTCR, were much shorter in this subgroup (28.7 months and 15 months, respectively) than in patients with sole TP53 disruption who did not reach the median OS (P<0.0001) and showed a TTCR of 30 months (P<0.0001) [28].
Similar observations were reported by Baliakas et al, who found a trend for a significantly shorter TFT in patients with del(17p) and concomitant CK as compared with patients with del(17p) alone (P=0.06) [23].
Finally, Le Bris et al, reported a dismal outcome in patients with concurrent TP53 disruption, U-IGHV and CK, with a shorter PFS (12 vs. 55 months; HR: 8.1, 95% CI 1.13–57.39; P<0.0001) and shorter OS (5–year OS: 21.3 ± 18% vs 84.7 ± 8%; HR: 25.7, 95% CI 1.15–574; P < 0.0001) as compared with the patients without this combination of adverse risk factors [29].
The CK may represent an unfavorable prognostic marker in the targeted therapy era
Whereas mechanism-based treatment with novel agents proved very efficacious in unfavorable genetic subsets of CLL (i.e. U-IGHV, TP53 disruption, 11q-) [79–81] several studies showed that the CK may have a negative prognostic role in R/R CLL receiving kinase targeted treatment or the BCL2 inhibitor venetoclax, as summarized in Table 2 [14, 61, 79, 82–83]. A retrospective analysis on 88 R/R CLL patients receiving Ibrutinib based regimens demonstrated that the CK was a stronger predictor of an inferior outcome than del(17p) [61]. A CK and del(17p) were found in 21/56 (38%) and 34/86 (40%) assessable cases, respectively, whereas they coexisted in 17 cases. At a median follow-up for surviving patients of 28 months (range, 14-48 months) the CK was associated with a shorter event free survival (EFS) (HR 6.6; 95% CI, 1.7-25.6; P = 0.006) and inferior OS (HR 5.9; 95% CI, 1.6-22.2; P = 0.008). Furthermore, a trend for a shorter EFS was observed in patients with a CK and del(17p) vs. those with sole del(17p) (P = 0.056), whereas no association was noted between del(17p) and OS (P = 0.885) [61]. It is worth noting that TP53 mutations were not assessed in this analysis. Brown et al, analysed the prognostic significance of CK in the phase 3 RESONATE study, including 195 R/R CLL patients treated with Ibrutinib [14]. At a median follow up of 19 months the presence of CK in 39/153 (25%) assessable patients did not show a significant impact on PFS (HR 1.53, 95% CI, 0.741-3.157; P=0.2476) and on OS (HR 1.86; 95% CI, 0.770-4.485; P=0.1610). It is noteworthy that, at a 5-year follow-up of phase-2 studies, the presence of CK was associated with a highly significant difference in median PFS in R/R CLL (31 months with CK compared to not reached in patients without CK and with 51 months in the entire cohort). However, the only genetic parameter retaining its adverse significance at multivariable analysis in this study was represented by the 17p deletion [79]. The CK and/or del(17p) may predispose to ibrutinib resistance through the development of the BTK C481S mutation or phospholipase Cγ2 (PLCγ2) activating mutation [61, 84–86].
Table 2. Impact of complex karyotype on R/R CLL patients treated with pathway inhibitors.
| Therapy and patient population | Tot. pts | CK | Median OS (months) |
OS | Median PFS (months) |
PFS | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| CK yes | CK no | HR (IC 95%) | P | CK yes | CK no | HR (IC 95%) | P | |||
| Ibrutinib in R/R CL [14, 61] | 88 | 21/56 (37.5%) |
25 | NR | 5.9 (1.6-22.2) |
0.008 | 19 | 39 | 6.6 (1.7-25.6)* |
0.006 |
| 195 | 39/153 (25.5%) |
NR | NR | 1.86 (0.770-4.485) |
0.161 | NR | NR | 1.53 (0.741-3.157) |
0.248 | |
| Idelalisib in R/R CLL [82] | 110 | 26/65 (40%) |
NR | NR | 1.78 (0.69-4.64) |
0.230 | 20.9 | 19.4 | 1.18 | 0.630 |
| Venetoclax in R/R CLL post KI [83] | 67 | 16/38 (42.1%) |
- | - | - | - | 16 | NR | 6.6 (1.5-29.8)** |
0.005 |
R/R relapsed/refractory; OS = overall survival; TFT = time to first treatment; EFS = event free survival; PFS = progression free survival; NR = not reached; *Event Free Surival (EFS); **Time to progression (TTP).
The CK was shown to have a strong adverse impact on outcome in patients treated with Venetoclax [83]. Among 67 R/R CLL patients treated with this BCL-2 inhibitor, 16 out of 38 (42%) assessable patients had CK. In univariate analysis the CK was associated with higher risk of progression (HR 6.6; 95% CI, 1.5-29.8; P = 0.005) along with fludarabine-refractory status (HR 7.01; 95%CI, 1.7-28.5; P = 0.002). Multivariate analysis was not performed due to the small sample size in this study. Interestingly, the presence of a CK increased the risk of progression among patients with F-refractory disease (P = 0.002), whereas TP53 mutation and/or del(17p) did not show any impact on time to progression (TTP) [83].
In patients treated with idelalisib and rituximab, the prognosis was not significantly influenced by the presence of a CK in the analysis by Kreuzer and coworkers, who reported their experience on 65 patients with available karyotype drawn from 110 R/R CLL patients receiving Idelalisib plus rituximab. With a relatively short follow-up (median of 21.4 months), 26 patients with CK showed no significant difference in terms of PFS and OS as compared with 39 patients without CK [82].
Prospective studies are needed to support the circumstantial evidence summarized here that a CK may represent a prognosticator in patients with R/R CLL receiving new oral agents.
CK and Richter transformation
The presence of CK was sporadically linked to the development of RT in previous reports [87–89]. In a retrospective study on CLL patients treated with first-line FCR, Le Bris et al, reported a CK in 1/4 cases with RT [29]. Anderson et al found a CK in 12 of 25 patients (48%) with progression on Venetoclax, including 8 of 17 patients with RT (47%) [83]. Rogers et al, studied the impact of CK in 46 CLL patients who developed RT. They reported a CK in 28/42 (67%) patients who subsequently developed RT and found that CK had an adverse impact at multivariate analysis on OS with the R-EPOCH regimen (HR 2.72; CI 95%, 1.14-6.52; P=0.025) [90].
In a recent analysis, Miller et al, found an association between near-tetraploidy (4 copies of most chromosomes within a cell) with CK and showed that 6/9 patients with this peculiar cytogenetic pattern developed RT. In a multivariate analysis near-terapolidy and CK represented independent predictors of ibrutinib discontinuation due to transformation [91].
Further studies are required to define the correlation between CK and RT
Perspectives
The importance of cytogenetic analysis in CLL was recognized by the 2018 guidelines of the International Workshop on CLL [92], which proposed the introduction of CBA in future prospective clinical trials to validate the prognostic and predictive value of karyotype aberrations [93]. The following issues represent an area of investigation which may facilitate the introduction of cytogenetic analysis in clinical practice.
Refinement of the definition of CK
Evidence was provided that this broad cytogenetic category, defined by the presence of at least 3 chromosome aberrations, should be regarded as heterogeneous. The patients with CK due to the coexistence of trisomy of chromosomes 12, 18 and 19, showed favourable clinicobiologic characteristics in terms of age (median 59 years), high incidence of mutated IGHV status and low frequency of TP53 disruption or NOTCH1 mutation (5% and 4% of cases, respectively) [23, 94, 95]. Likewise, Baliakas and colleagues identified a subset of patients with CK carrying +12, +19 plus other numerical and/or structural chromosome abnormalities (12% of the cases), which displayed indolent clinical course independent of clinical stage, IGHV mutational status and TP53 status [96]. These findings suggest that the cytogenetic complexity defined solely by numerical aberrations should not be regarded as an unfavorable prognostic marker in CLL. Recently, Rigolin et al, showed that within patients carrying CK as defined by the presence of 3 or more aberrations, the presence of unbalanced translocations (i.e. chromosome additions, derivatives, insertions, duplications, ring-, dicentric- and marker-chromosomes) was associated with a worse outcome in terms of OS and TTFT (HR 2.773; 95% CI, 1.056-7.281; P=0.038 and HR 2.375; 95% CI, 1.027-5.492; P=0.043) [97]. Interestingly, a distinct mRNA expression profile, with a deregulation of genes involved in cell cycle control and DNA damage response, was documented in patients with a CK carrying unbalanced rearrangements [97].
Finally, of the presence of ≥5 chromosomal aberrations, referred to as high-CK, predicted for a particularly aggressive clinical course in a large multicentre study, possibly due to a strong association with TP53 disruptions (P<0.001) [23]. More recently high-CK (i.e. ≥5 chromosomal aberrations) was shown to represent a strong adverse prognosticator independent of clinical stage, IGHV mutational status and TP53 status [96].
New methods of detection
CBA is somewhat laborious, has a low sensitivity and requires mitotic stimulation of fresh or frozen living cells. Hence, alternative methods of detection of genomic complexity were developed. The array-based (CGH/SNP) analysis offers the opportunity to study the CLL genome, does not require in vitro mitogens and allows for the detection of subtle DNA gains and losses [98]. Leeksma et al, recently analysed 1911 patients with monoclonal B-cell lymphocytosis and treatment-naïve CLL and found that 451/1911 cases (24%) displayed genome complexity (defined as the presence of ≥3 structural and/or numerical aberrations) [99]. This study also showed that array-analysis detected more aberrations than CBA (2.35 vs 1.84, 95% CI paired differences 0.221-0.798) [99].
Diagnostic platforms using whole genome sequencing (WGS) to detect single nucleotide variants and insertion/deletions are being developed and validated for potential usage in clinical practice [100]. Although these methods will likely provide comprehensive genomic characterisation of CLL and will represent alternative method to recognize the prognostic or predictive role genetic lesions in trials, they still require standardization and a univocal definition of “genome complexity”.
At the moment CBA represents a standardized tool for risk assessment in CLL, providing complementary information to FISH and traditional genetic studies of recurrent mutations. Because cytogenetic laboratories are available in the majority of hematologic centres and the mitotic yield greatly improved with novel mitogens, CBA could be incorporated in prospective trials to definitely establish its predictive power in an era in which both CIT and new mechanism-based treatment are available [92].
SUPPLEMENTARY MATERIALS FIGURES AND TABLES
ACKNOWLEDGMENTS AND FUNDING
This work was supported by AIL Ferrara, Beat Leukemia Foundation, Milan (www.beat-leukemia.org/uk/index.asp), the Fondo di Ateneo per la Ricerca 2013, 2014, 2016, 2017 of the University of Ferrara (G.M.R., A.C.), Fondi incentivazione alla ricerca 2017 (FIR) of the University of Ferrara to GMR, Programma Ricerca Regione Università 2007-2009 University of Ferrara (G.M.R., A.C.), Programmi di Ricerca di Rilevante Interesse Nazionale (PRIN) 2008 (A.C.), Ricerca Finalizzata (A.C.; project RF-2011-02349712), Ministero dell’Istruzione, dell’Università e della Ricerca PRIN 2015 (A.C.; project 2015ZMRFEA), E.V. and E.S. are supported by AIL Ferrara.
Author contributions
All authors edited and approved the final manuscript.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.The Surveillance Epidemiology and End Results (SEER) Program of the National Cancer Institute Cancer fact sheets: chronic lymphocytic leukemia (CLL) https://seer.cancer.gov/statfacts/html/clyl.html.
- 3.Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352:804–815. doi: 10.1056/NEJMra041720. [DOI] [PubMed] [Google Scholar]
- 4.Cuneo A, Cavazzini F, Ciccone M, Daghia G, Sofritti O, Saccenti E, Negrini M, Rigolin GM. Modern treatment in chronic lymphocytic leukemia: impact on survival and efficacy in high-risk subgroups. Cancer Med. 2014;3:555–564. doi: 10.1002/cam4.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zenz T, Fröhling S, Mertens D, Döhner H, Stilgenbauer S. Moving from prognostic to predictive factors in chronic lymphocytic leukaemia (CLL) Best Pract Res Clin Haematol. 2010;23:71–84. doi: 10.1016/j.beha.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 6.Oscier DG, Thompsett A, Zhu D, Stevenson FK. Differential rates of somatic hypermutation in V(H) genes among subsets of chronic lymphocytic leukemia defined by chromosomal abnormalities. Blood. 1997;89:4153–4160. [PubMed] [Google Scholar]
- 7.Döhner H, Stilgenbauer S, Benner A, Leupolt E, Kröber A, Bullinger L, Döhner K, Bentz M, Lichter P. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 8.Zenz T, Kröber A, Scherer K, Häbe S, Bühler A, Benner A, Denzel T, Winkler D, Edelmann J, Schwänen C, Döhner H, Stilgenbauer S. Monoallelic TP53 inactivation is associated with poor prognosis in chronic lymphocytic leukemia: results from a detailed genetic characterization with long-term follow-up. Blood. 2008;112:3322–3329. doi: 10.1182/blood-2008-04-154070. [DOI] [PubMed] [Google Scholar]
- 9.Rosenquist R, Cortese D, Bhoi S, Mansouri L, Gunnarsson R. Prognostic markers and their clinical applicability in chronic lymphocytic leukemia: where do we stand? Leuk Lymphoma. 2013;54:2351–2364. doi: 10.3109/10428194.2013.783913. [DOI] [PubMed] [Google Scholar]
- 10.International CLL-IPI working group An international prognostic index for patients with chronic lymphocytic leukaemia (CLL-IPI): a meta-analysis of individual patient data. Lancet Oncol. 2016;17:779–790. doi: 10.1016/S1470-2045(16)30029-8. [DOI] [PubMed] [Google Scholar]
- 11.Byrd JC, Furman RR, Coutre SE, Flinn IW, Burger JA, Blum KA, Grant B, Sharman JP, Coleman M, Wierda WG, Jones JA, Zhao W, Heerema NA, et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med. 2013;369:32–42. doi: 10.1056/NEJMoa1215637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furman RR, Sharman JP, Coutre SE, Cheson BD, Pagel JM, Hillmen P, Barrientos JC, Zelenetz AD, Kipps TJ, Flinn I, Ghia P, Eradat H, Ervin T, et al. Idelalisib and rituximab in relapsed chronic lymphocytic leukemia. N Engl J Med. 2014;370:997–1007. doi: 10.1056/NEJMoa1315226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberts AW, Davids MS, Pagel JM, Kahl BS, Puvvada SD, Gerecitano JF, Kipps TJ, Anderson MA, Brown JR, Gressick L, Wong S, Dunbar M, Zhu M, et al. Targeting BCL2 with Venetoclax in Relapsed Chronic Lymphocytic Leukemia. N Engl J Med. 2016;374:311–322. doi: 10.1056/NEJMoa1513257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown JR, Hillmen P, O'Brien S, Barrientos JC, Reddy NM, Coutre SE, Tam CS, Mulligan SP, Jaeger U, Barr PM, Furman RR, Kipps TJ, Cymbalista F, et al. Extended follow-up and impact of high-risk prognostic factors from the phase 3 RESONATE study in patients with previously treated CLL/SLL. Leukemia. 2018;32:83–91. doi: 10.1038/leu.2017.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharman JP, Coutre SE, Furman RR, Cheson BD, Pagel JM, Hillmen P, Barrientos JC, Zelenetz AD, Kipps TJ, Flinn IW, Ghia P, Hallek M, Coiffier B, et al. Second Interim Analysis of a Phase 3 Study of Idelalisib (ZYDELIG®) Plus Rituximab (R) for Relapsed Chronic Lymphocytic Leukemia (CLL): Efficacy Analysis in Patient Subpopulations with Del(17p) and Other Adverse Prognostic Factors. Blood. 2014;124:330. [Google Scholar]
- 16.Stilgenbauer S, Eichhorst B, Schetelig J, Coutre S, Seymour JF, Munir T, Puvvada SD, Wendtner CM, Roberts AW, Jurczak W, Mulligan SP, Böttcher S, Mobasher M, et al. Venetoclax in relapsed or refractory chronic lymphocytic leukaemia with 17p deletion: a multicentre, open-label, phase 2 study. Lancet Oncol. 2016;17:768–778. doi: 10.1016/S1470-2045(16)30019-5. [DOI] [PubMed] [Google Scholar]
- 17.Robèrt KH, Möller E, Gahrton G, Eriksson H, Nilsson B. B-cell activation of peripheral blood lymphocytes from patients with chronic lymphatic leukaemia. Clin Exp Immunol. 1978;33:302–308. [PMC free article] [PubMed] [Google Scholar]
- 18.Juliusson G, Oscier DG, Fitchett M, Ross FM, Stockdill G, Mackie MJ, Parker AC, Castoldi GL, Cuneo A, Knuutila S, Elonen E, Gahrton G. Prognostic subgroups in B-cell chronic lymphocytic leukemia defined by specific chromosomal abnormalities. N Engl J Med. 1990;323:720–724. doi: 10.1056/NEJM199009133231105. [DOI] [PubMed] [Google Scholar]
- 19.Gahrton G, Juliusson G, Robèrt KH, Friberg K. Role of chromosomal abnormalities in chronic lymphocytic leukemia. Blood Rev. 1987;1:183–92. doi: 10.1016/0268-960x(87)90034-8. [DOI] [PubMed] [Google Scholar]
- 20.Mayr C, Speicher MR, Kofler DM, Buhmann R, Strehl J, Busch R, Hallek M, Wendtner CM. Chromosomal translocations are associated with poor prognosis in chronic lymphocytic leukemia. Blood. 2006;107:742–751. doi: 10.1182/blood-2005-05-2093. [DOI] [PubMed] [Google Scholar]
- 21.Bardi A, Cavazzini F, Rigolin GM, Tammiso E, Volta E, Pezzolo E, Formigaro L, Sofritti O, Daghia G, Ambrosio C, Rizzotto L, Abass AE, D'Auria F, et al. Employment of oligodeoxynucleotide plus interleukin-2 improves cytogenetic analysis in splenic marginal zone lymphoma. J Biomed Biotechnol. 2011;2011:691493. doi: 10.1155/2011/691493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haferlach C, Dicker F, Schnittger S, Kern W, Haferlach T. Comprehensive genetic characterization of CLL: a study on 506 cases analysed with chromosome banding analysis, interphase FISH, IgV(H) status and immunophenotyping. Leukemia. 2007;21:2442–2451. doi: 10.1038/sj.leu.2404935. [DOI] [PubMed] [Google Scholar]
- 23.Baliakas P, Iskas M, Gardiner A, Davis Z, Plevova K, Nguyen-Khac F, Malcikova J, Anagnostopoulos A, Glide S, Mould S, Stepanovska K, Brejcha M, Belessi C, et al. Chromosomal translocations and karyotype complexity in chronic lymphocytic leukemia: a systematic reappraisal of classic cytogenetic data. Am J Hematol. 2014;89:249–255. doi: 10.1002/ajh.23618. [DOI] [PubMed] [Google Scholar]
- 24.Herling CD, Klaumünzer M, Rocha CK, Altmüller J, Thiele H, Bahlo J, Kluth S, Crispatzu G, Herling M, Schiller J, Engelke A, Tausch E, Döhner H, et al. Complex karyotypes and KRAS and POT1 mutations impact outcome in CLL after chlorambucil-based chemotherapy or chemoimmunotherapy. Blood. 2016;128:395–404. doi: 10.1182/blood-2016-01-691550. [DOI] [PubMed] [Google Scholar]
- 25.Rigolin GM, Cibien F, Martinelli S, Formigaro L, Rizzotto L, Tammiso E, Saccenti E, Bardi A, Cavazzini F, Ciccone M, Nichele I, Pizzolo G, Zaja F, et al. Chromosome aberrations detected by conventional karyotyping using novel mitogens in chronic lymphocytic leukemia with “normal” FISH: correlations with clinicobiologic parameters. Blood. 2012;119:2310–2313. doi: 10.1182/blood-2011-11-395269. [DOI] [PubMed] [Google Scholar]
- 26.Rigolin GM, Saccenti E, Bassi C, Lupini L, Quaglia FM, Cavallari M, Martinelli S, Formigaro L, Lista E, Bardi MA, Volta E, Tammiso E, Melandri A, et al. Extensive next-generation sequencing analysis in chronic lymphocytic leukemia at diagnosis: clinical and biological correlations. J Hematol Oncol. 2016;9:88. doi: 10.1186/s13045-016-0320-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rigolin GM, Cavallari M, Quaglia FM, Formigaro L, Lista E, Urso A, Guardalben E, Liberatore C, Faraci D, Saccenti E, Bassi C, Lupini L, Bardi MA, et al. comorbidities and the complex karyotype are associated with an inferior outcome independently of CLL-IPI. Blood. 2017;129:3495–3498. doi: 10.1182/blood-2017-03-772285. [DOI] [PubMed] [Google Scholar]
- 28.Rigolin GM, Formigaro L, Cavallari M, Quaglia FM, Lista E, Urso A, Guardalben E, Martinelli S, Saccenti E, Bassi C, Lupini L, Bardi MA, Volta E, et al. An extensive molecular cytogenetic characterization in high-risk chronic lymphocytic leukemia identifies karyotype aberrations and TP53 disruption as predictors of outcome and chemorefractoriness. Oncotarget. 2017;8:28008–28020. doi: 10.18632/oncotarget.15883. https://doi.org/10.18632/oncotarget.15883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Bris Y, Struski S, Guièze R, Rouvellat C, Prade N, Troussard X, Tournilhac O, Béné MC, Delabesse E, Ysebaert L. Major prognostic value of complex karyotype in addition to TP53 and IGHV mutational status in first-line chronic lymphocytic leukemia. Hematol Oncol. 2017;35:664–670. doi: 10.1002/hon.2349. [DOI] [PubMed] [Google Scholar]
- 30.Mato AR, Hill BT, Lamanna N, Barr PM, Ujjani CS, Brander DM, Howlett C, Skarbnik AP, Cheson BD, Zent CS, Pu JJ, Kiselev P, Foon K, et al. Optimal sequencing of ibrutinib, idelalisib, and venetoclax in chronic lymphocytic leukemia: results from a multicenter study of 683 patients. Ann Oncol. 2017;28:1050–1056. doi: 10.1093/annonc/mdx031. [DOI] [PubMed] [Google Scholar]
- 31.Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T, Dombret H, Ebert BL, Fenaux P, Larson RA, Levine RL, Lo-Coco F, Naoe T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strickland SA, Sun Z, Ketterling RP, Cherry AM, Cripe LD, Dewald G, Fernandez HF, Hicks GA, Higgins RR, Lazarus HM, Litzow MR, Luger SM, Paietta EM, et al. ECOG-ACRIN Cancer Research Group Independent Prognostic Significance of Monosomy 17 and Impact of Karyotype Complexity in Monosomal Karyotype/Complex Karyotype Acute Myeloid Leukemia: Results from Four ECOG-ACRIN Prospective Therapeutic Trials. Leuk Res. 2017;59:55–64. doi: 10.1016/j.leukres.2017.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greenberg PL, Tuechler H, Schanz J, Sanz G, Garcia-Manero G, Solé F, Bennett JM, Bowen D, Fenaux P, Dreyfus F, Kantarjian H, Kuendgen A, Levis A, et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood. 2012;120:2454–2465. doi: 10.1182/blood-2012-03-420489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valcárcel D, Ademà V, Solé F, Ortega M, Nomdedeu B, Sanz G, Luño E, Cañizo C, de la Serna J, Ardanaz M, Marco V, Collado R, Grau J, et al. Complex, not monosomal, karyotype is the cytogenetic marker of poorest prognosis in patients with primar. J Clin Oncol. 2011;29:392–397. doi: 10.1200/JCO.2012.41.6073. [DOI] [PubMed] [Google Scholar]
- 35.Gangat N, Caramazza D, Vaidya R, George G, Begna K, Schwager S, Van Dyke D, Hanson C, Wu W, Pardanani A, Cervantes F, Passamonti F, Tefferi A. DIPSS plus: a refined Dynamic International Prognostic Scoring System for primary myelofibrosis that incorporates prognostic information from karyotype, platelet count, and transfusion status. J Clin Oncol. 2011;29:392–397. doi: 10.1200/JCO.2010.32.2446. [DOI] [PubMed] [Google Scholar]
- 36.Tefferi A, Jimma T, Gangat N, Vaidya R, Begna KH, Hanson CA, Van Dyke DL, Caramazza D, Pardanani A. Predictors of greater than 80% 2-year mortality in primary myelofibrosis: a Mayo Clinic study of 884 karyotypically annotated patients. Blood. 2011;118:4595–4598. doi: 10.1182/blood-2011-08-371096. [DOI] [PubMed] [Google Scholar]
- 37.Juliusson G, Robèrt KH, Ost A, Friberg K, Biberfeld P, Nilsson B, Zech L, Gahrton G. Prognostic information from cytogenetic analysis in chronic B-lymphocytic leukemia and leukemic immunocytoma. Blood. 1985;65:134–141. [PubMed] [Google Scholar]
- 38.Haferlach C, Dicker F, Weiss T, Schnittger S, Beck C, Grote-Metke A, Oruzio D, Kerm W, Haferlach T. Toward a comprehensive prognostic scoring system in chronic lymphocytic leukemia based on a combination of genetic parameters. Genes Chromosomes Cancer. 2010;49:851–859. doi: 10.1002/gcc.20794. [DOI] [PubMed] [Google Scholar]
- 39.Rigolin GM, del Giudice I, Formigaro L, Saccenti E, Martinelli S, Cavallari M, Lista E, Tammiso E, Volta E, Lupini L, Bassi C, Bardi A, Sofritti O, et al. Chromosome aberrations detected by conventional karyotyping using novel mitogens in chronic lymphocytic leukemia: Clinical and biologic correlations. Genes Chromosomes Cancer. 2015;54:818–826. doi: 10.1002/gcc.22293. [DOI] [PubMed] [Google Scholar]
- 40.Chiorazzi N. Implications of new prognostic markers in chronic lymphocytic leukemia. Hematology Am Soc Hematol Educ Program. 2012;2012:76–87. doi: 10.1182/asheducation-2012.1.76. [DOI] [PubMed] [Google Scholar]
- 41.Martinelli S, Cuneo A, Formigaro L, Cavallari M, Lista E, Quaglia FM, Ciccone M, Bardi A, Volta E, Tammiso E, Saccenti E, Sofritti O, Daghia G, et al. Identifying High-Risk Chronic Lymphocytic Leukemia: A Pathogenesis-Oriented Appraisal of Prognostic and Predictive Factors in Patients Treated with Chemotherapy with or without Immunotherapy. Mediterr J Hematol Infect Dis. 2016;8:e2016047. doi: 10.4084/MJHID.2016.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burns A, Alsolami R, Becq J, Stamatopoulos B, Timbs A, Bruce D, Robbe P, Vavoulis D, Clifford R, Cabes M, Dreau H, Taylor J, Knight SJL, et al. Whole-genome sequencing of chronic lymphocytic leukaemia reveals distinct differences in the mutational landscape between IgHVmut and IgHVunmut subgroups. Leukemia. 2018;32:332–342. doi: 10.1038/leu.2017.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guièze R, Wu CJ. Genomic and epigenomic heterogeneity in chronic lymphocytic leukemia. Blood. 2015;126:445–453. doi: 10.1182/blood-2015-02-585042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomay K, Fedder C, Hofmann W, Kreipe H, Stadler M, Titgemeyer J, Zander I, Schlegelberger B, Göhring G. Telomere shortening, TP53 mutations and deletions in chronic lymphocytic leukemia result in increased chromosomal instability and breakpoint clustering in heterochromatic regions. Ann Hematol. 2017;96:1493–1500. doi: 10.1007/s00277-017-3055-1. [DOI] [PubMed] [Google Scholar]
- 45.Lazarian G, Tausch E, Eclache V, Sebaa A, Bianchi V, Letestu R, Collon JF, Lefebvre V, Gardano L, Varin-Blank N, Soussi T, Stilgenbauer S, Cymbalista F, et al. TP53 mutations are early events in chronic lymphocytic leukemia disease progression and precede evolution to complex karyotypes. Int J Cancer. 2016;139:1759–1763. doi: 10.1002/ijc.30222. [DOI] [PubMed] [Google Scholar]
- 46.Roos G, Kröber A, Grabowski P, Kienle D, Bühler A, Döhner H, Rosenquist R, Stilgenbauer S. Short telomeres are associated with genetic complexity, high-risk genomic aberrations, and short survival in chronic lymphocytic leukemia. Blood. 2008;111:2246–2252. doi: 10.1182/blood-2007-05-092759. [DOI] [PubMed] [Google Scholar]
- 47.Mansouri L, Grabowski P, Degerman S, Svenson U, Gunnarsson R, Cahill N, Smedby KE, Geisler C, Juliusson G, Roos G, Rosenquist R. Short telomere length is associated with NOTCH1/SF3B1/TP53 aberrations and poor outcome in newly diagnosed chronic lymphocytic leukemia patients. Am J Hematol. 2013;88:647–651. doi: 10.1002/ajh.23466. [DOI] [PubMed] [Google Scholar]
- 48.Steinbrecher D, Jebaraj BMC, Schneider C, Edelmann J, Cymbalista F, Leblond V, Delmer A, Ibach S, Tausch E, Scheffold A, Bloehdorn J, Hallek M, Dreger P, et al. Telomere length in poor-risk chronic lymphocytic leukemia: associations with disease characteristics and outcome. Leuk Lymphoma. 2018;59:1614–1623. doi: 10.1080/10428194.2017.1390236. [DOI] [PubMed] [Google Scholar]
- 49.Deng Y, Chan SS, Chang S. Telomere dysfunction and tumour suppression: the senescence connection. Nat Rev Cancer. 2008;8:450–458. doi: 10.1038/nrc2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brugat T, Nguyen-Khac F, Grelier A, Merle-Béral H, Delic J. Telomere dysfunction-induced foci arise with the onset of telomeric deletions and complex chromosomal aberrations in resistant chronic lymphocytic leukemia cells. Blood. 2010;116:239–249. doi: 10.1182/blood-2009-12-257618. [DOI] [PubMed] [Google Scholar]
- 51.Landau DA, Tausch E, Taylor-Weiner AN, Stewart C, Reiter JG, Bahlo J, Kluth S, Bozic I, Lawrence M, Böttcher S, Carter SL, Cibulskis K, Mertens D, et al. Mutations driving CLL and their evolution in progression and relapse. Nature. 2015;526:525–530. doi: 10.1038/nature15395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 53.Siu KT, Xu Y, Swartz KL, Bhattacharyya M, Gurbuxani S, Hua Y, Minella AC. Chromosome instability underlies hematopoietic stem cell dysfunction and lymphoid neoplasia associated with impaired Fbw7-mediated cyclin E regulation. Mol Cell Biol. 2014;34:3244–3258. doi: 10.1128/MCB.01528-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coste I, Le Corf K, Kfoury A, Hmitou I, Druillennec S, Hainaut P, Eychene A, Lebecque S, Renno T. Dual function of MyD88 in RAS signaling and inflammation, leading to mouse and human cell transformation. J Clin Invest. 2010;120:3663–3667. doi: 10.1172/JCI42771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oliveira-Santos W, Rabello DA, Lucena-Araujo AR, de Oliveira FM, Rego EM, Pittella Silva F, Saldanha-Araujo F. Residual expression of SMYD2 and SMYD3 is associated with the acquisition of complex karyotype in chronic lymphocytic leukemia. Tumour Biol. 2016;37:9473–9481. doi: 10.1007/s13277-016-4846-z. [DOI] [PubMed] [Google Scholar]
- 56.Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, Kubicek S, Opravil S, Jenuwein T, Berger SL. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444:629–632. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- 57.Cho HS, Hayami S, Toyokawa G, Maejima K, Yamane Y, Suzuki T, Dohmae N, Kogure M, Kang D, Neal DE, Ponder BA, Yamaue H, Nakamura Y, et al. RB1 methylation by SMYD2 enhances cell cycle progression through an increase of RB1 phosphorylation. Neoplasia. 2012;14:476–486. doi: 10.1593/neo.12656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang G, Mercado-Uribe I, Multani AS, Sen S, IeM Shih, Wong KK, Gershenson DM, Liu J. RAS promotes tumorigenesis through genomic instability induced by imbalanced expression of Aurora-A and BRCA2 in midbody during cytokinesis. Int J Cancer. 2013;133:275–85. doi: 10.1002/ijc.28032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lucena-Araujo AR, de Oliveira FM, Leite-Cueva SD, dos Santos GA, Falcao RP, Rego EM. High expression of AURKA and AURKB is associated with unfavorable cytogenetic abnormalities and high white blood cell count in patients with acute myeloid leukemia. Leuk Res. 2011;35:260–264. doi: 10.1016/j.leukres.2010.07.034. [DOI] [PubMed] [Google Scholar]
- 60.Pflug N, Bahlo J, Shanafelt TD, Eichhorst BF, Bergmann MA, Elter T, Bauer K, Malchau G, Rabe KG, Stilgenbauer S, Döhner H, Jäger U, Eckart MJ, et al. Development of a comprehensive prognostic index for patients with chronic lymphocytic leukemia. Blood. 2014;124:49–62. doi: 10.1182/blood-2014-02-556399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thompson PA, O'Brien SM, Wierda WG, Ferrajoli A, Stingo F, Smith SC, Burger JA, Estrov Z, Jain N, Kantarjian HM, Keating MJ. Complex karyotype is a stronger predictor than del(17p) for an inferior outcome in relapsed or refractory chronic lymphocytic leukemia patients treated with ibrutinib-based regimens. Cancer. 2015;121:3612–3621. doi: 10.1002/cncr.29566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.González-Gascón y, Marín I, Hernández-Sanchez M, Rodríguez-Vicente AE, Puiggros A, Collado R, Luño E, González T, Ruiz-Xivillé N, Ortega M, Gimeno E, Muñoz C, Infante MS, Delgado J, et al. Grupo Español de Leucemia Linfática Crónica (GELLC) and Grupo Cooperativo Español de Citogenética Hematológica (GCECGH) Characterizing patients with multiple chromosomal aberrations detected by FISH in chronich lymphocytic leukemia. Leuk Lymphoma. 2018;59:633–642. doi: 10.1080/10428194.2017.1349901. [DOI] [PubMed] [Google Scholar]
- 63.Puiggros A, Collado R, Calasanz MJ, Ortega M, Ruiz-Xivillé N, Rivas-Delgado A, Luño E, González T, Navarro B, García-Malo M, Valiente A, Hernández JA, Ardanaz MT, et al. Patients with chronic lymphocytic leukemia and complex karyotype show an adverse outcome even in absence of TP53/ATM FISH deletions. Oncotarget. 2017;8:54297–54303. doi: 10.18632/oncotarget.17350. https://doi.org/10.18632/oncotarget.17350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gaidano G, Rossi D. The mutational landscape of chronic lymphocytic leukemia and its impact on prognosis and treatment. Hematology Am Soc Hematol Educ Program. 2017;1:329–337. doi: 10.1182/asheducation-2017.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nadeu F, Delgado J, Royo C, Baumann T, Stankovic T, Pinyol M, Jares P, Navarro A, Martín-García D, Beà S, Salaverria I, Oldreive C, Aymerich M, et al. Clinical impact of clonal and subclonal TP53, SF3B1, BIRC3, NOTCH1, and ATM mutations in chronic lymphocytic leukemia. Blood. 2016;127:2122–2130. doi: 10.1182/blood-2015-07-659144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kastenhuber ER, Lowe SW. Putting p53 in Context. Cell. 2017;170:1062–1078. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Blackford AN, Jackson SP. ATM, ATR, DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol Cell. 2017;66:801–817. doi: 10.1016/j.molcel.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 68.Gattei V, Bulian P, Del Principe MI, Zucchetto A, Maurillo L, Buccisano F, Bomben R, Dal-Bo M, Luciano F, Rossi FM, Degan M, Amadori S, Del Poeta G. Relevance of CD49d protein expression as overall survival and progressive disease prognosticator in chronic lymphocytic leukemia. Blood. 2008;111:865–873. doi: 10.1182/blood-2007-05-092486. [DOI] [PubMed] [Google Scholar]
- 69.Geisler CH, Philip P, Hansen MM. B-cell chronic lymphocytic leukaemia: clonal chromosome abnormalities and prognosis in 89 cases. Eur J Haematol. 1989;43:397–403. doi: 10.1111/j.1600-0609.1989.tb00326.x. [DOI] [PubMed] [Google Scholar]
- 70.Juliusson G, Gahrton G, Chromosome aberrations in B-cell chronic lymphocytic leukemia Pathogenetic and clinical implications. Cancer Genet Cytogenet. 1990;45:143–160. doi: 10.1016/0165-4608(90)90079-p. [DOI] [PubMed] [Google Scholar]
- 71.Oscier DG, Stevens J, Hamblin TJ, Pickering RM, Lambert R, Fitchett M. Correlation of chromosome abnormalities with laboratory features and clinical course in B-cell chronic lymphocytic leukaemia. Br J Haematol. 1990;76:352–358. doi: 10.1111/j.1365-2141.1990.tb06367.x. [DOI] [PubMed] [Google Scholar]
- 72.Travella A, Ripollés L, Aventin A, Rodríguez A, Bezares RF, Caballín MR, Slavutsky I, Structural alterations in chronic lymphocytic leukaemia Cytogenetic and FISH analysis. Hematol Oncol. 2013;31:79–87. doi: 10.1002/hon.2025. [DOI] [PubMed] [Google Scholar]
- 73.Badoux XC, Keating MJ, Wang X, O'Brien SM, Ferrajoli A, Faderl S, Burger J, Koller C, Lerner S, Kantarjian H, Wierda WG. Fludarabine, cyclophosphamide, and rituximab chemoimmunotherapy is highly effective treatment for relapsed patients with CLL. Blood. 2011;117:3016–3024. doi: 10.1182/blood-2010-08-304683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Badoux XC, Keating MJ, Wang X, O'Brien SM, Ferrajoli A, Faderl S, Burger J, Koller C, Lerner S, Kantarjian H, Wierda WG. Cyclophosphamide, fludarabine, alemtuzumab, and rituximab as salvage therapy for heavily pretreated patients with chronic lymphocytic leukemia. Blood. 2011;118:2085–2093. doi: 10.1182/blood-2011-03-341032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Buhmann R, Kurzeder C, Rehklau J, Westhaus D, Bursch S, Hiddemann W, Haferlach T, Hallek M, Schoch C. CD40L stimulation enhances the ability of conventional metaphase cytogenetics to detect chromosome aberrations in B-cell chronic lymphocytic leukaemia cells. Br J Haematol. 2002;118:968–975. doi: 10.1046/j.1365-2141.2002.03719.x. [DOI] [PubMed] [Google Scholar]
- 76.Foà R, Del Giudice I, Cuneo A, Del Poeta G, Ciolli S, Di Raimondo F, Lauria F, Cencini E, Rigolin GM, Cortelezzi A, Nobile F, Callea V, Brugiatelli M, et al. Chlorambucil plus rituximab with or without maintenance rituximab as first-line treatment for elderly chronic lymphocytic leukemia patients. Am J Hematol. 2014;89:480–486. doi: 10.1002/ajh.23668. [DOI] [PubMed] [Google Scholar]
- 77.Takahashi K, Hu B, Wang F, Yan Y, Kim E, Vitale C, Patel KP, Strati P, Gumbs C, Little L, Tippen S, Song X, Zhang J, et al. Clinical implications of cancer gene mutations in patients with chronic lymphocytic leukemia treated with lenalidomide. Blood. 2018;131:1820–1832. doi: 10.1182/blood-2017-11-817296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Blanco G, Puiggros A, Baliakas P, Athanasiadou A, García-Malo M, Collado R, Xochelli A, Rodríguez-Rivera M, Ortega M, Calasanz MJ, Luño E, Vargas M, Grau J, et al. Karyotypic complexity rather than chromosome 8 abnormalities aggravates the outcome of chronic lymphocytic leukemia patients with TP53 aberrations. Oncotarget. 2016;7:80916–80924. doi: 10.18632/oncotarget.13106. https://doi.org/10.18632/oncotarget.13106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.O'Brien S, Furman RR, Coutre S, Flinn IW, Burger JA, Blum K, Sharman J, Wierda W, Jones J, Zhao W, Heerema NA, Johnson AJ, Luan Y, et al. Single-Agent Ibrutinib in Treatment-Naïve and Relapsed/Refractory Chronic Lymphocytic Leukemia: A 5-Year Experience. Blood. 2018;131:1910–1919. doi: 10.1182/blood-2017-10-810044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jones JA, Robak T, Brown JR, Awan FT, Badoux X, Coutre S, Loscertales J, Taylor K, Vandenberghe E, Wach M, Wagner-Johnston N, Ysebaert L, Dreiling L, et al. Efficacy and safety of idelalisib in combination with ofatumumab for previously treated chronic lymphocytic leukaemia: an open-label, randomised phase 3 trial. Lancet Haematol. 2017;4:e114–e126. doi: 10.1016/S2352-3026(17)30019-4. [DOI] [PubMed] [Google Scholar]
- 81.Seymour JF, Kipps TJ, Eichhorst B, Hillmen P, D'Rozario J, Assouline S, Owen C, Gerecitano J, Robak T, De la Serna J, Jaeger U, Cartron G, Montillo M, et al. Venetoclax- Rituximab in Relapsed or Refractory Chronic Lymphocytic Leukemia. N Engl J Med. 2018;378:1107–1120. doi: 10.1056/NEJMoa1713976. [DOI] [PubMed] [Google Scholar]
- 82.Kreuzer KA, Furman R, Stilgenbauer S, Dubowy RL, Kim Y, Munugalavadla V, Lilienweiss E, Reinhardt H, Pettitt A, Hallek M. Outcome of Patients with Complex Karyotype in a Phase 3 Randomized Study of Idelalisib Plus Rituximab for Relapsed Chronic Lymphocytic Leukemia. Blood. 2016;128:192. [Google Scholar]
- 83.Anderson MA, Tam C, Lew TE, Juneja S, Juneja M, Westerman D, Wall M, Lade S, Gorelik A, Huang DCS, Seymour JF, Roberts AW. Clinicopathological features and outcomes of progression of CLL on the BCL2 inhibitor venetoclax. Blood. 2017;129:3362–3370. doi: 10.1182/blood-2017-01-763003. [DOI] [PubMed] [Google Scholar]
- 84.Jain P, Keating M, Wierda W, Estrov Z, Ferrajoli A, Jain N, George B, James D, Kantarjian H, Burger J, O'Brien S. Outcomes of patients with chronic lymphocytic leukemia after discontinuing ibrutinib. Blood. 2015;125:2062–2067. doi: 10.1182/blood-2014-09-603670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Woyach JA, Furman RR, Liu TM, Ozer HG, Zapatka M, Ruppert AS, Xue L, Li DH, Steggerda SM, Versele M, Dave SS, Zhang J, Yilmaz AS, et al. Resistance mechanisms for the Bruton's tyrosine kinase inhibitor ibrutinib. N Engl J Med. 2014;370:2286–94. doi: 10.1056/NEJMoa1400029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, Sougnez C, Stewart C, Sivachenko A, Wang L, Wan Y, Zhang W, Shukla SA, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hébert J, Jonveaux P, d'Agay MF, Berger R. Cytogenetic studies in patients with Richter's syndrome. Cancer Genet Cytogenet. 1994;73:65–68. doi: 10.1016/0165-4608(94)90184-8. [DOI] [PubMed] [Google Scholar]
- 88.Arranz E, Martínez B, Richart A, Echezarreta G, Román A, Rivas C, Benítez J. Increased C-MYC oncogene copy number detected with combined modified comparative genomic hybridization and FISH analysis in a Richter syndrome case with complex karyotype. Cancer Genet Cytogenet. 1998;106:80–83. doi: 10.1016/s0165-4608(98)00037-5. [DOI] [PubMed] [Google Scholar]
- 89.Woroniecka R, Rymkiewicz G, Grygalewicz B, Błachnio K, Rygier J, Jarmuż-Szymczak M, Ratajczak B, Pieńkowska-Grela B. Cytogenetic and flow cytometry evaluation of Richter syndrome reveals MYC, CDKN2A, IGH alterations with loss of CD52, CD62L and increase of CD71 antigen expression as the most frequent recurrent abnormalities. Am J Clin Pathol. 2015;143:25–35. doi: 10.1309/AJCPATRQWANW2O3N. [DOI] [PubMed] [Google Scholar]
- 90.Rogers KA, Huang Y, Ruppert AS, Salem G, Stephens DM, Heerema NA, Andritsos LA, Awan FT, Byrd JC, Flynn JM, Maddocks KJ, Jones JA. A single-institution retrospective cohort study of first-line R-EPOCH chemoimmunotherapy for Richter syndrome demonstrating complex chronic lymphocytic leukaemia karyotype as an adverse prognostic factor. Br J Haematol. 2018;180:259–266. doi: 10.1111/bjh.15035. [DOI] [PubMed] [Google Scholar]
- 91.Miller CR, Ruppert AS, Heerema NA, Maddocks KJ, Labanowska J, Breidenbach H, Lozanski G, Zhao W, Gordon AL, Jones JA, Flynn JM, Jaglowski SM, Andritsos LA, et al. Near-tetraploidy is associated with Richter transformation in chronic lymphocytic leukemia patients receiving ibrutinib. Blood Adv. 2017;1:1584–1588. doi: 10.1182/bloodadvances.2017007302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Döhner H, Hillmen P, Keating M, Montserrat E, Chiorazzi N, Stilgenbauer S, Rai KR, Byrd JC, et al. Guidelines for diagnosis, indications for treatment, response assessment and supportive management of chronic lymphocytic leukemia. Blood. 2018;131:2745–2760. doi: 10.1182/blood-2017-09-806398. [DOI] [PubMed] [Google Scholar]
- 93.Hallek M, Shanafelt TD, Eichhorst B. Chronic lymphocytic leukaemia. Lancet. 2018;391:1524–1537. doi: 10.1016/S0140-6736(18)30422-7. [DOI] [PubMed] [Google Scholar]
- 94.Ibbotson R, Athanasiadou A, Sutton LA, Davis Z, Gardiner A, Baliakas P, Gunnarsson R, Anagnostopoulos A, Juliusson G, Rosenquist R, Oscier D, Stamatopoulos K. Coexistence of trisomies of chromosomes 12 and 19 in chronic lymphocytic leukemia occurs exclusively in the rare IgG-positive variant. Leukemia. 2012;26:170–172. doi: 10.1038/leu.2011.186. [DOI] [PubMed] [Google Scholar]
- 95.Baliakas P, Puiggros A, Xochelli A, Sutton LA, Nguyen-Khac F, Gardiner A, Plevova K, Minga E, Hadzidimitriou A, Walewska R, McCarthy H, Ortega M, Collado R, et al. Additional trisomies amongst patients with chronic lymphocytic leukemia carrying trisomy 12: the accompanying chromosome makes a difference. Haematologica. 2016;101:e299–302. doi: 10.3324/haematol.2015.140202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Baliakas P, Jeromin S, Iskas M, Puiggros A, Plevova K, Xochelli A, Delgado J, Kotaskova J, Stalika E, Abrisqueta P, Durechova K, Papaioannou G, Collado R, et al. Cytogenetic complexity in chronic lymphocytic leukemia: definitions, associations with other biomarkers and clinical impact; a retrospective study on behalf of ERIC. EHA Learning Center. 2017. p. 181748. (Abstract release date: May 18, 2017)
- 97.Rigolin GM, Saccenti E, Guardalben E, Cavallari M, Formigaro L, Zagatti B, Visentin A, Mauro FR, Lista E, Bassi C, Lupini L, Quaglia FM, Urso A, et al. In chronic lymphocytic leukaemia with complex karyotype, major structural abnormalities identify a subset of patients with inferior outcome and distinct biologic characteristics. Br J Haematol. 2018;181:229–233. doi: 10.1111/bjh.15174. [DOI] [PubMed] [Google Scholar]
- 98.Braggio E, Fonseca R, Kay NE. CGH protocols: chronic lymphocytic leukemia. Methods Mol Biol. 2013;973:87–98. doi: 10.1007/978-1-62703-281-0_6. [DOI] [PubMed] [Google Scholar]
- 99.Leeksma A, Baliakas P, Mellink C, Moysiadis T, Puiggros A, Plevova K, van den Berg E, Eldering E, Skaftason A, Mansouri L, Gunnarsson R, Ghia P, Espinet B, et al. Genomic Complexity in Chronic Lymphocytic Leukemia Defined By Array-Based Analysis: Definitions, Associations with Other Biomarkers and Clinical Impact. Blood. 2017;130:4280. [Google Scholar]
- 100.Klintman J, Barmpouti K, Knight SJL, Robbe P, Dreau H, Clifford R, Ridout K, Burns A, Timbs A, Bruce D, Antoniou P, Sosinsky A, Becq J, et al. Clinical-grade validation of whole genome sequencing reveals robust detection of low-frequency variants and copy number alterations in CLL. Br J Haematol. 2018. May 29, [Epub ahead of print] [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.