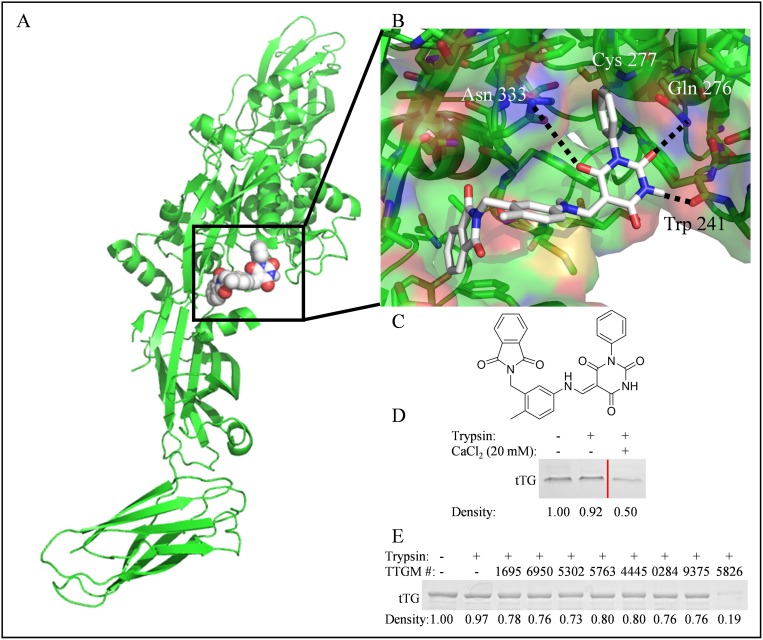
Figure 2. The identification of TTGM 5826 as a potential modulator of tTG conformation.
(A) For screening purposes, molecules were docked to the substrate binding site (boxed) of the open state of tTG (green, pdb ID: 2Q3Z). Shown is TTGM 5826 (space filling, white) docked to tTG. (B) Enlarged view of TTGM 5826 (white) docked to the crosslinking active site of tTG (green, pdb ID: 2Q3Z). The phenyl ring of TTGM 5826 projects into a deep pocket formed around the catalytic Cys 277, while the barbiturate ring is predicted to engage in hydrogen bonds with Trp 241, Gln 276, and Asn 333. The flexible linker allows the molecule to wrap around a ‘hump’ in the binding site, while the phthalamide projects into a second deep hydrophobic pocket. (C) The chemical structure of TTGM 5826 is composed of four moieties: a phenyl ring (lower left), a barbiturate (upper left), a tolyl ring (upper right), and a phthalamide (lower right). (D) tTG was incubated on ice with or without 20 mM CaCl2 for 5 minutes, and then each sample was incubated without (lane 1) or with trypsin (lanes 2 & 3) for 3 hours. The samples were resolved by SDS-PAGE and stained with Coomassie blue. Lighter bands indicate that a greater amount of tTG was digested. Lanes of samples where lower concentrations of CaCl2 were used were spliced out for clarity, and are indicated by the red line. (E) tTG was incubated on ice with the indicated small molecule (TTGM #), or DMSO, for 5 minutes, at which point trypsin was added, as indicated, and the reactions were processed as described in (C). TTGM molecules are labeled by the last four digits of their ChemBridge catalog number. Densiometric quantitation was performed with ImageJ. Band densities are reported as fractional density of the trypsin-free control band.