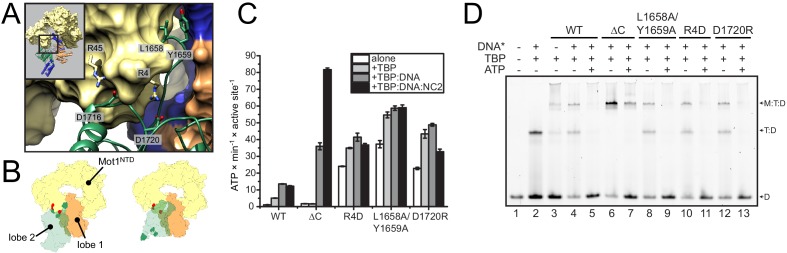
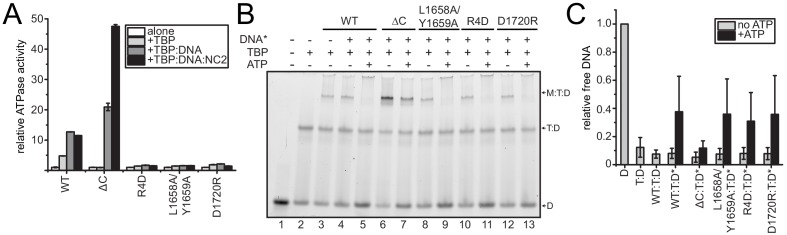
Figure 2. Analysis of CtMot1 mutants.
(A) View at the interface between RecA2A (green cartoon), HR1/2 (yellow surface), and lobe 1 (orange/blue surface). Residues analyzed in this study are shown as sticks and labelled accordingly. (B) Cartoon model showing the positions of mutations in the Mot1NTD (red spheres on yellow surface) and in lobe 2 (green spheres on green surface). Left: orientation as in the CtMot1 crystal structure. Right: CtMot1 with ATPase modeled as in the S. cerevisiae Chd1:nucleosome complex, that is the ATP hydrolysis-competent conformation (Farnung et al., 2017). (C) ATPase activity of the mutants. Error bars represent standard deviations from three technical replicates. CtMot1WT is labelled as WT, CtMot1ΔC as ΔC. (D) Electrophoretic mobility shift assay showing ATP-dependent dissociation of Mot1:TBP:DNA and TBP:DNA complexes. All CtMot1 constructs form ternary complexes with labelled DNA and TBP (M:T:D). In the presence of ATP and unlabeled competitor DNA (DNA*), wild-type CtMot1 (WT), L1658/Y1659, R4D, and D1720R mutants fully disrupt M:T:D and T:D complexes (lanes 5, 9, 11, and 13, respectively), whereas CtMot1ΔC (ΔC) is less efficient (lane 7).